Interactive predictive toxicology with Bioclipse and OpenTox
Abstract:
Computational predictive toxicology draws knowledge from many independent sources, providing a rich support tool to assess a wide variety of toxicological properties. A key example would be for it to complement alternative testing methods. The integration of Bioclipse and OpenTox permits toxicity prediction based on the analysis of chemical structures, and visualization of the substructure contributions to the toxicity prediction. In analogy of the decision support that is already in use in the pharmaceutical industry for designing new drug leads, we use this approach in two case studies in malaria research, using a combination of local and remote predictive models. This way, we find drug leads without predicted toxicity.
2.1 Introduction
Much is already known about the toxicity of small molecules, but yet it is a challenge to predict the toxicology of new compounds. At the same time, society demands insight into the toxicity of compounds, such as that outlined by the REACH regulations in the European Union [1]. Where and when experimental testing of toxicity is expensive or unnecessary, experimental data may under certain conditions be complemented by alternative methods. Among these alternative methods are computational tools, such as read-across and (Q)SAR, that can predict a wide variety of toxicological properties. A good coverage of predictions with different alternative models can help provide reasonable estimations for the toxicity of a compound.
Such predictive toxicology draws knowledge from many independent sources, which need to be integrated to provide a weight of evidence on the toxicity of untested chemical compounds. Typical sources include in vivo and in vitro experimental databases such as ToxCast [2] and SuperToxic [3], literature-derived databases, such as SIDER summarizing adverse reactions [4], and also computational resources based on toxicity data for other compounds including DSSTox [5]. And somehow all this information should be aggregated and presented to the user in such a way that the various, potentially contradictory, pieces of information can help reach a weighted decision on the toxicity of the compound.
Visualization of this information is, therefore, an important tool, and should preferably be linked to the chemical structure of the compound itself. Further visualization that will be important is that of relevant life science data, such as gene, protein and biological pathway information [6–8] or metabolic reactions [9]. Bioclipse was designed to provide such interactive data analysis for the life sciences, although the resources are not yet as tightly integrated as other sources.
Underlying this knowledge integration, there must be a platform that allows researchers to use multiple prediction services. This is exactly what the recently introduced Bioclipse–OpenTox platform is providing. This chapter will describe how this platform can be used to interactively study the toxicity of chemical structures using computational toxicology tools. Its ability to interactively predict toxicity and the ability to dynamically discover computational services, allows users to get the latest insights into toxicity predictions while hiding technicalities. Other tools that provide similar functionality include the OECD QSAR ToolBox and ToxTree [12, 13]. Bioclipse, however, is not a platform targeted at toxicity alone, and has been used for other scientific fields too, taking advantage of the ease by which other functionality, both local and remote, can be integrated [14–16].
This chapter will not go into too much technical detail, but instead provide two example use cases of the platform. Detailed descriptions can be found in the papers describing the OpenTox Application Programming Interface (API) [17] and the AMBIT implementation of this API [18], the second Bioclipse paper describing the scriptability [19], a recently published description of the integration of these tools [10], and the paper describing the technologies used to glue together the components [20]. For the user of the platform it suffices to know that all functionality of the Bioclipse-OpenTox interoperability is accessible both graphically and by means of scripting. Table 2.1 shows an overview of all currently available functionality.
Table 2.1
Bioclipse-OpenTox functionality from the Graphical User Interface is also available from the scripting environment. Scriptable commands (left column) and descriptions (right column) for various groups of functionality are provided in this table
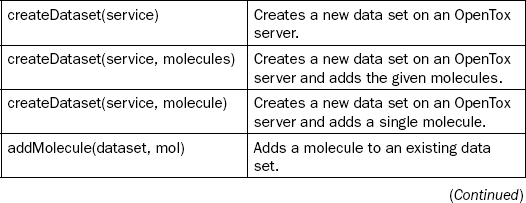
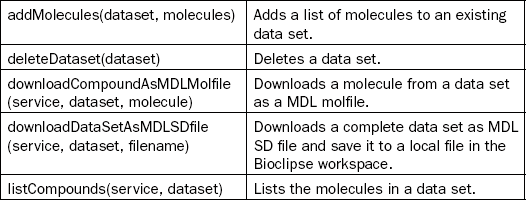
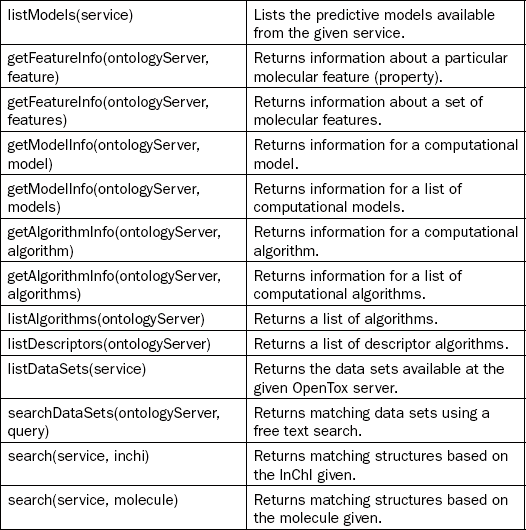
This chapter will first outline a few basic tasks one can perform with the Bioclipse–OpenTox platform, including the calculation of molecular descriptors (which can also be used to calculate properties important to toxicology, including logP and pKa), the sharing of data on toxins and toxicants online, and how Bioclipse supports authorization and authentication on the OpenTox network. After that, two more elaborate use cases will be presented, which will make use of the Decision Support extension. Bioclipse Decision Support was developed by the Department of Pharmaceutical Biosciences, Uppsala University, Sweden, and AstraZeneca R&D, Molndal, Sweden, with the goal of building an extensible platform for integrating multiple predictive models with a responsive user interface. Bioclipse Decision Support is now being used for ADME-T predictions within AstraZeneca R&D.
2.2 Basic Bioclipse-OpenTox interaction examples
As an introduction to the Bioclipse-OpenTox interoperability, this chapter will first introduce a few short examples. Of course, to reproduce these examples yourself, you may have to familiarize yourself with the Bioclipse software, which takes, like any other software, some time to learn. However, the examples given in this chapter will not be that basic. For more tutorials on how to use Bioclipse, we refer the reader to http://www.opentox.org/tutorials/bioclipse.
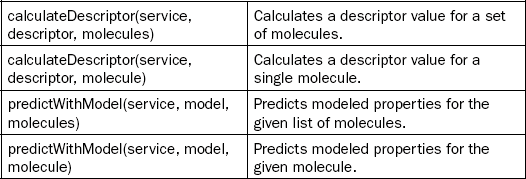
Figure 2.1 shows the first Bioclipse–OpenTox integration and highlights the Bioclipse QSAR environment for calculating molecular properties and theoretical descriptors [21]. This environment has been extended to discover OpenTox services on the internet. Using this approach, Bioclipse has access to the most recent descriptors relevant to toxicity predictions. The figure shows the equivalence of a number of descriptors provided by both local services (CDK and CDK REST, provided by the Chemistry Development Kit library [22]) and OpenTox, as well as the Ionization Potential descriptor provided only via an OpenTox computational service. Practically, these online computational services are found by querying a registry of OpenTox services: this registry makes use of the OpenTox ontology [17], which Bioclipse queries, as outlined in [10]. The ability to compute descriptors using various local and remote providers creates a flexible application for integration of numerical inputs for statistical modeling of toxicologically relevant endpoints, as well as comparison of various predictive models for a more balanced property analysis.
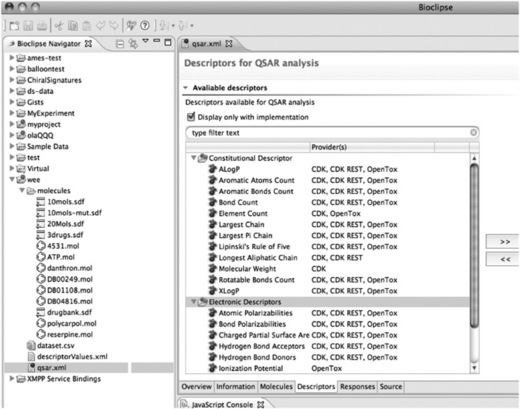
Figure 2.1 Integration of online OpenTox descriptor calculation services in the Bioclipse QSAR environment. Molecular descriptors are frequently used in computational toxicology models. This screenshot from Bioclipse QSAR shows descriptors discovered on the Internet (providers: OpenTox and CDK REST) in combination with local software (provider: CDK)
A second basic example is the sharing of data on the OpenTox network, which demonstrates how all Graphical User Interface (GUI) interaction in Bioclipse can also be performed using a scripting language. Bioclipse extends scripting languages like JavaScript and Groovy with life sciences-specific extensions, defining the Bioclipse Scripting Language (BSL). These extensions are available for much open source life sciences software, including Jmol, the Chemistry Development Kit, but also the OpenTox API. The latter is used in, for example, Figure 2.2, which shows the Bioclipse dialog for uploading a small data set with ten neurotoxin structures to an OpenTox server. This dialog asks to which OpenTox server the structures should be uploaded (the Ambit2 server is selected, http://apps.ideaconsult.net:8080/ambit2/), a title under which this data set will be available (Ten neurotoxins found in Wikipedia’), and the data license under which the data will be available to others (the Creative Commons Zero license in this case [23]).
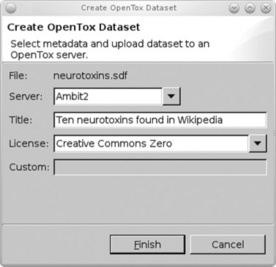
Figure 2.2 The Bioclipse Graphical User Interface for uploading data to OpenTox. Sharing new toxicological data about molecular structures can be done by uploading the data to an OpenTox server. This Bioclipse dialog shows a selected MDL SD file with ten neurotoxins (neurotoxins.sdf) being shared on the Ambit2 server, the OpenTox server to upload to, providing a title for the data set, and the CC0 waiver (see main text). Clicking the Finish button will upload the structures and open a web browser window in Bioclipse with the resulting online data set (see Figure 2.3)
At a scripting level, this dialog makes use of the createDataSet(service, molecules), setDatasetLicense(datasetURI, licenseURI), and set DatasetTitle(datasetURI, title) commands, as listed in Table 2.1. The latter two methods use the data set’s Universal Resource Identifier (URI) returned by the first method. The dialog, however, opens the data set’s web page in a browser window in Bioclipse (see Figure 2.3) when the upload has finished. This is currently not possible using the scripting language.
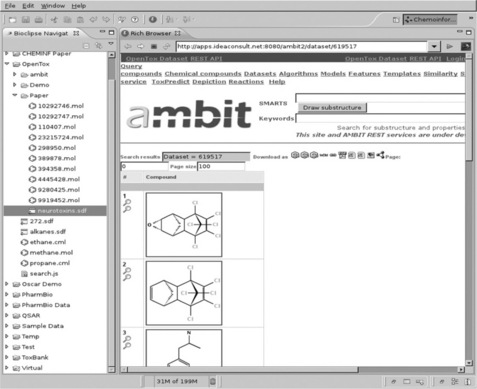
Figure 2.3 OpenTox web page showing uploaded data. Screenshot of Bioclipse showing a web browser window with the neurotoxins data hosted on the Ambit2 OpenTox server after the upload, as shown in Figure 2.2 (see http://apps.ideaconsult.net:8080/ambit2/ dataset/619517)
Most of the other Bioclipse-OpenTox integration takes advantage of the fact that Bioclipse has all GUI functionality matched by a scripted equivalent too. The use of the BSL directly allows interaction with the OpenTox network to be automated, combined with other Bioclipse functionality into larger workflows, and makes it easier to share procedures with others using social scientific sites like MyExperiment [24].
For example, a BSL script equivalent for the earlier descriptor calculation GUI would look as follows:
This script combines functionality from the Chemistry Development Kit plug-in for Bioclipse via the cdk BSL extension with the OpenTox functionality provided by the opentox extension.
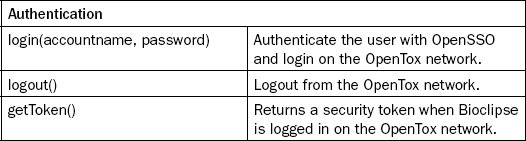
The third basic functionality of the Bioclipse-OpenTox interoperability we highlight here is the support for accessing protected resources within the OpenTox network. Despite preferences of the authors, we acknowledge that not all scientific data will be Open Data in the foreseeable future. As such, authentication and authorization (A&A) are important features of data access. OpenTox implements both aspects, and provides web services for A&A, allowing users to log in and out of OpenTox applications, accompanied by policy-based specification of OpenTox resource access permissions. Additionally, the same mechanism is used to restrict access to calculation procedures, allowing exposure of software with commercial licenses as protected OpenTox resources.
Bioclipse was extended to support the authentication functionality, allowing OpenTox servers to properly authorize user access to particular web services and data sets. The OpenTox account information is registered with Bioclipse’s keyring system, centralizing logging in and out onto remote services, providing the GUI for adding a new OpenTox account and to log in and out. The corresponding script commands for the authentication are given in Authentication category in Table 2.1. Support of authentication is important for industrial environments in which confidential data are handled.
With these basics covered, we will now turn to two use cases much more interesting from a toxicology perspective. For these, we will use the Tres Cantos Antimalarial Compound Set, TCAMS – a collection of 13 533 compounds of the Tres Cantos Medicines Development Campus of GlaxoSmithKline deposited in 2010 at the ChEMBL Neglected Tropical Disease Database (https://www.ebi.ac.uk/chemblntd), making the data publicly available [25]. These compounds will be tested against a number of toxicity-related predictive models, as outlined in Table 2.2, see [16] for a more detailed description. The Decision Support plug-in allows detailed information as to how the decision for a particular endpoint was reached, using a variety of data types outlined in Table 2.3. The OpenTox extension currently does not make use of this, as there is no ontology available at this moment to communicate this information. However, this is under development.
Table 2.2
Description of the local endpoints provided by the default Bioclipse Decision Support extension. The OpenTox integrates further tests, which are not described in this table. An up-to-date overview of services available on the OpenTox network is provided at http://apps.ideaconsult.net:8080/ToxPredict#Models
| Ames mutagenicity | The Ames Salmonella microsome mutagenicity assay (AMES test) indicates if a compound can induce mutations to DNA. |
| CPDB | Carcinogenic Potency Database (CPDB) contains on compounds known to lead to cancer. |
| AhR | Aryl hydrocarbon receptor (AhR) is a transcription factor involved in the regulation of xenobioticmetabolizing enzymes, such as cytochrome P450. |
Table 2.3
Various data types are used by the various predictive models described in Table 2.2 to provide detailed information about what aspects of the molecules contributed to the decision on the toxicity
| Structural alerts | A substructure that has been associated with an alert, in our case a chemical liability. Can be implemented in many ways, most common is using SMARTS patterns. Also referred to as toxicophore if alerting for toxicity. |
| Signature alerts | A type of structural alert implemented using Signatures [26]. |
| Signature significance | A QSAR model which is capable of, apart from a prediction, to return the most significant signature in the prediction [27]. |
| Exact match | An identical chemical structure was found in the training data. Can be implemented by InChI and Molecular Signature. |
| Near Neighbor match | A similar chemical structure was found in the training data. Commonly implemented by binary |
Each of the chemicals in the TCAMS inhibits growth of the 3D7 strain of Plasmodium falciparum – the malaria causing parasite – by at least 80% at a concentration of 2 p,M. Many also show a similar effect against the multidrug-resistant P. falciparum strain DD2. Evidence for liver toxicity is provided by growth inhibition data against human hepatoma HepG2 cells. Of all TCAMS compounds, 857 are annotated with a so-called target hypothesis. This target hypothesis had been obtained by comparing each compound with public and GSK-internal data of compound-target relationships, accepting a target hypothesis if a homologue to the identified target exists in P. falciparum and unless many compound-target relationships had been identified for a given chemical. Of this subset, 233 are annotated as potential Ser/Thr kinase inhibitors.
2.3 Use Case 1: Removing toxicity without interfering with pharmacology
From the TCAMS data set, we select the compound TCMDC-135308 for further investigation. TCMDC-135308 is similar (Tanimoto = 0.915) to quinazoline 3d (both shown in Table 2.4), a potent human-TGF-81 inhibitor (Transforming Growth Factor-β1). TCMDC-135308 inhibits growth of the P. falciparum strain 3D7 by 98% at a concentration of 2 μM (XC50 = 700 nM). It is also active against the multidrug-resistant P. falciparum strain DD2, inhibiting its growth by 63% at a concentration of 2 μM. The compound has not shown significant growth inhibition of human HepG2 cells (5% at a concentration of 10 μM).
Table 2.4
Structures created from SMILES representations with the Bioclipse New from SMILES wizard for various structures discussed in the use cases
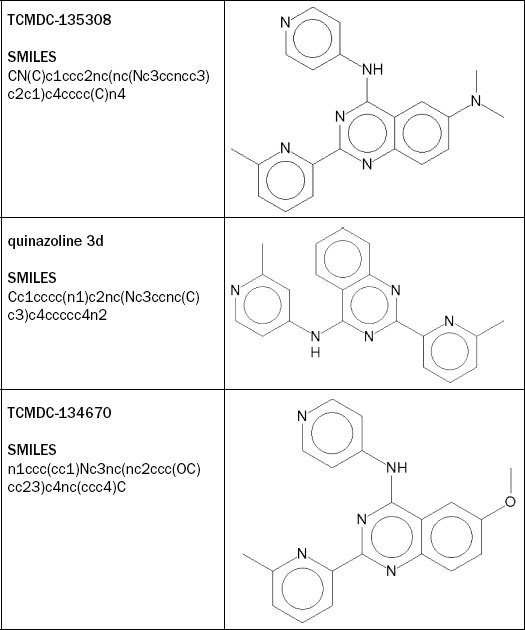
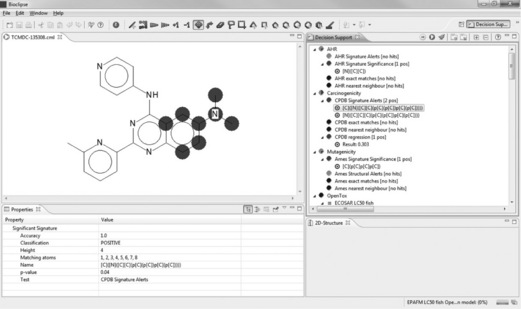
We import TCMDC-135308 using its SMILES to Bioclipse, log in to OpenTox and move to the Decision Support Perspective, where we run all models. It turns out TCMDC-135308 contains a CPDB Signature Alert for Carcinogenicity (see Figure 2.4). Signature Alerts in Bioclipse Decision Support are discriminative signatures identified according to [8], which can be visualized as substructures in the chemical structure of the query molecule.
The carcinogenicity prediction is supported by some positive predictions with OpenTox models ‘IST DSSTox Carcinogenic Potency DBS Mouse’, ‘IST DSSTox Carcinogenic Potency DBS SingleCellCall’ and ‘ToxTree: Benigni/Bossa rules for carcinogenicity and mutagenicity’ (structural alert for genotoxic carcinogenicity) as shown in Figure 2.5. Also, the OpenTox models ‘IST DSSTox Carcinogenic Potency DBS Mutagenicity’, ‘IST Kazius-Bursi Salmonella Mutagenicity’ and ‘ToxTree: Structure Alerts for the in vivo micronucleus assay in rodents’ are positive.
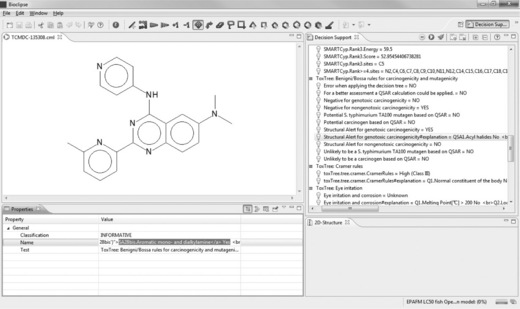
Figure 2.5 Identification of the structural alert in the ToxTree Benigni/Bossa model for carcinogenicity and mutagenicity, available via OpenTax
To identify the structural alerts, we click on the ‘Structural Alert for genotoxic carcinogenicity#explanation’ line in the Decision Support window. In the property tab, the property ‘Name’ lists the contents of the #explanation obtained from OpenTox. One can either scroll along the explanation in the value field of the ‘Name’ property, or copy and paste its content to a text editor to identify which structural alert was fired. For our compound of interest, it is the presence of a dialkylamine, which overlaps with the CPDB Signature Alert for Carcinogenicity. The presence of a dialkylamine is also a structural alert for the ‘in vivo micronucleus assay in rodents’ model.
Analyzing the crystal structure of the inhibitor quinazoline 3d bound to human-TGF-β1 (PDB entry 3HMM [29]), we find that the dimethylamine is not deeply buried in the binding pocket, but rather exposed, without making crucial interactions. This is visualized with the Jmol viewer in Bioclipse in Figure 2.6. The dimethylamino group might therefore be easily replaceable without markedly affecting the activity of the compound.
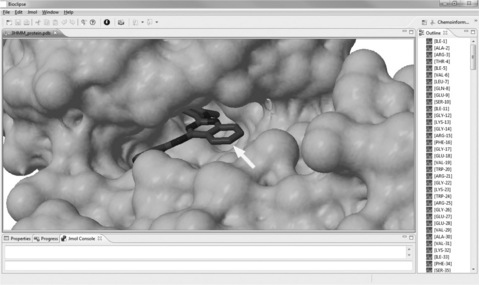
Figure 2.6 Crystal structure of human TGF-β1 with the inhibitor quinazoline 3d bound (PDB-entry 3HMM). The dimethylamino group of TCMDC-135308 is bound to the carbon atom of quinazoline 3d highlighted by the white arrow, thus pointing toward the water-exposed side of the binding pocket. The image was generated from the 3HMM crystal structure, hiding all water molecules, using the Jmol editor in Bioclipse
However, as we do not know the exact target protein in P. falciparum, we cannot exclude that the dimethylamino group is important for the activity or selectivity of the compound. Therefore, we try to replace it with a bioisosteric group, the simplest one being a methoxy group.
In the Bioclipse compound window, we modify the structure of TCMDC-135308 by renaming the nitrogen atom of the dimethyl amino group to oxygen, and deleting one of the methyl groups of the former dimethyl amino group.
Re-running all predictions, we find that there are no CPDP signature alerts anymore, and that the OpenTox Benigni/Bossa rules predict the modified compound to be negative for both genotoxic and non-genotoxic carcinogenicity (Figure 2.7). Also the ‘IST DSSTox Carcinogenic Potency DBS Mutagenicity’ and ‘IST DSSTox Carcinogenic Potency DBS Mouse’ models are now negative. The OpenTox model for the in vivo micronucleus assay is still positive because of the presence of two hydrogen-bond acceptors that are three bonds apart. Also the OpenTox ‘IST DSSTox Carcinogenic Potency DBS SingleCellCall’ and ‘IST Kazius-Bursi Salmonella mutagenicity’ models are still positive. In addition, the ‘IST DSSTox Carcinogenic Potency DBS MultiCellCall’ model is now positive. As expected, physico-chemical properties such as pKa or XLogP are not, or only marginally, affected (no change to pKa, XLogP changes from 0.845 to 0.551), and also, no new positive toxicity predictions are obtained due to the change.
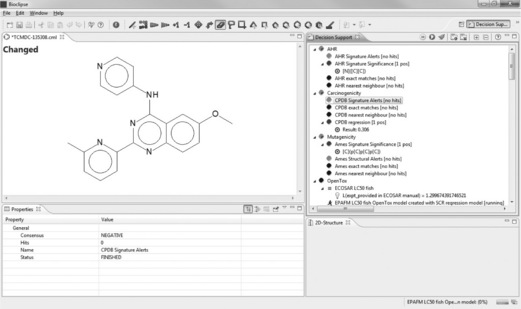
Figure 2.7 Replacing the dimethylamino group of TCMDC-135308 with a methoxy group resolves the CPDB signature alert as well as the ToxTree Benigni/Bossa Structure Alerts for carcinogenicity and mutagenicity as provided by OpenTox. The image illustrates the absence of CPDB signature alerts
Thus, our bioisosteric replacement of the dimethylamino group has resolved several toxicity predictions, while hopefully only marginally affecting the compound’s activity. As it turns out, the methoxy variant we constructed based on TCMDC-135308 exists in TCAMS as well. It is TCMDC-134670. Its activity against P. falciparum 3D7 is almost identical to that of TCMDC-135308 (101% growth inhibition at 2 μM, XC50 = 670 nM), whereas its activity against P. falciparum DD2 is slightly decreased (from 63% to 42% growth inhibition at 2 μM). TCMDC-134670 is also less active in the in vitro hepatotoxicity assay reported (− 1% growth inhibition of human HepG2 cells at a concentration of 10 μM, compared to 5% obtained with TCMDC-135308).
2.4 Use Case 2: Toxicity prediction on compound collections
As a second, illustrative example, we look at the US FDA’s Adverse Events Reporting System database (AERS) [30]. This database is a unique source of in vivo data on observations of the adverse outcomes of human toxicities of drugs. Pharmatrope [31] has processed the AERS data according to statistical considerations, and created the Titanium Adverse Events Database and Models. For all compounds in the TCAMS we predicted association with groups of human adverse events related to the hepatobiliary tract and classified the compounds’ association with these adverse events.
All data on the annotated kinase inhibitors in TCAMS – namely the compounds’ identifiers and SMILES, the inhibition of P. falciparum 3D7 and DD2, of human HepG2 cells, and the degree of association with human adverse events – was combined into an SDF file and imported into Bioclipse. Double-clicking the imported file in the Bioclipse Navigator opens the table, which allows the rearrangement of the columns (Figure 2.8).
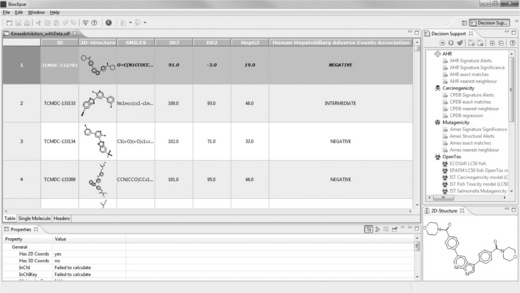
Figure 2.8 Annotated kinase inhibitors of the TCAMS, imported into Bioclipse as SDF together with data on the association with human adverse events
The imported SDF file contained a field for the compounds’ SMILES. However, we will not be using the SMILES here, so we hide the column by right-clicking on its header and selecting ‘Hide Column’. Clicking again on a column header, all compounds are selected. Right-clicking and selecting ‘Decision Support’ in the menu that opens gives the list of Bioclipse toxicity predictions that can be run on the compounds (Figure 2.9). We select the CPDB Signature Alerts.
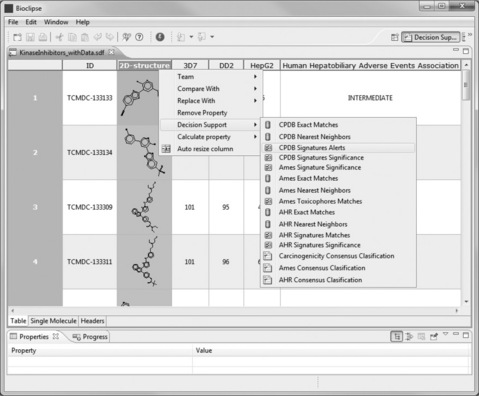
Figure 2.9 Applying toxicity models to sets of compounds from within the Bioclipse Molecule TableEditor
While running the calculations, Bioclipse displays a progress bar, so that users can monitor how far the calculation has proceeded. In a similar manner we calculate the Ames Signature Significance and the AHR Signature Significance. Once the computations are completed, we can save the table under a new name.
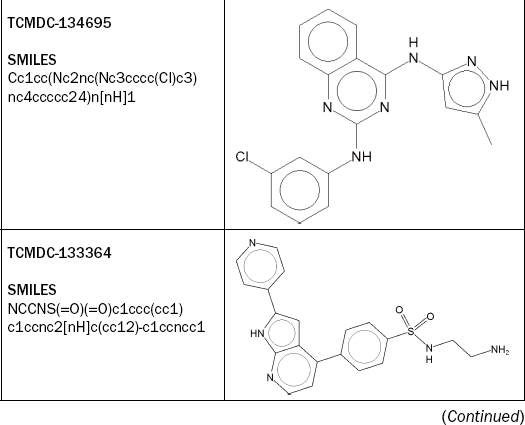
Browsing the table, we can identify some interesting compounds, for example TCMDC-133364 is very active against both P. falciparum strains, without inhibiting the growth of the HepG2 cells. The compound’s association with human adverse events is negative, and three out of four Bioclipse models are negative as well (Figure 2.10). Double-clicking on the 2D-structure diagram of TCMDC-133364 opens the compound in the Single Molecule View. We can now change to the Decision Support perspective and apply the OpenTox models on the selected compound (Figure 2.11).
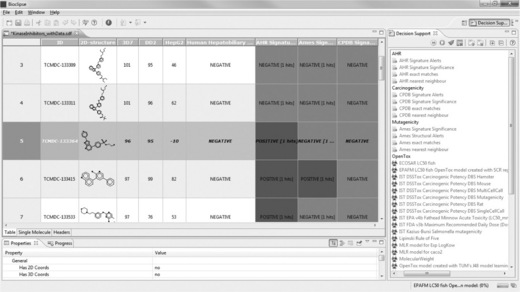
Figure 2.10 Adding Decision Support columns to the molecule table. The highlighted compound – TCMDC-133364 – is an interesting candidate as it is highly active against both strains of P falciparum while being inactive against human HepG2 cells. Also, the compound is predicted negative for three out of four toxicity models included
The positive AHR Signature Significance can be traced back to individual atoms in the Decision Support View, allowing for judgment of the prediction’s relevance. The negative prediction for carcinogenicity by the CPDB model in Bioclipse is confirmed by the negative predictions of the OpenTox models ‘IST DSSTox Carcinogenic Potency DBS Hamster’, ‘IST DSSTox Carcinogenic Potency DBS Mouse’ and ‘ToxTree: Benigni/Bossa rules for carcinogenicity and mutagenicity’. Only the OpenTox models ‘1ST DSSTox Carcinogenic Potency DBS MultiCellCall’ and ‘IST DSSTox Carcinogenic Potency DBS SingleCellCall’ are positive. For mutagenicity we have the negative prediction by Bioclipse’s Ames model, the negative prediction with the OpenTox Benigni/Bossa model and the ‘IST Kazius-Bursi Salmonella mutagenicity’ model. However, we obtain a positive prediction with the OpenTox ‘ToxTree: Structure Alerts for the in vivo micronucleus assay’ model. Inspecting the details of that prediction, we see that it is related to hydrogen-bond acceptors separated by three covalent bonds in the sulfonamide part of the molecule.
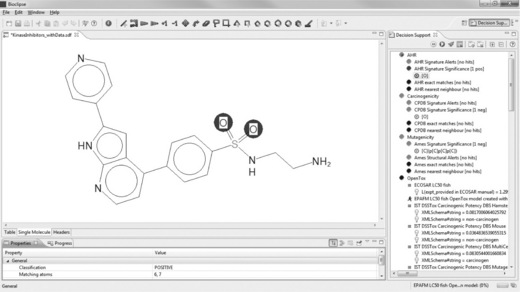
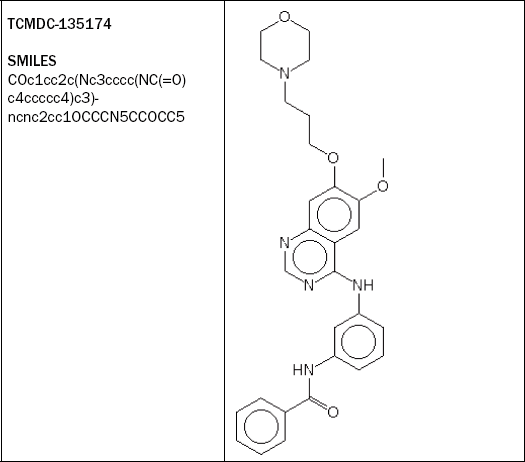
TCMDC-135174 is also very active against both P. falciparum strains, without inhibiting the growth of the HepG2 cells, and its association with human adverse events, as well as the Ames Toxicophore Match and the AHR Signature Match, are also negative. However, the Bioclipse CPDB Signature Match is positive, as is visible in Figure 2.12.
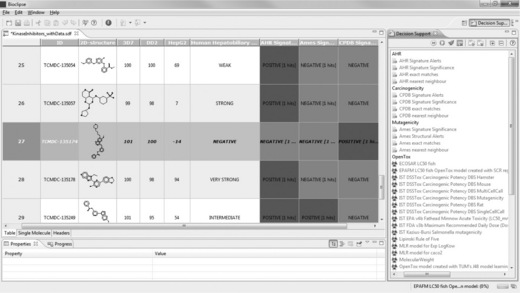
Figure 2.12 The highlighted compound – TCMDC-135174 (row 27) – is an interesting candidate as it is highly active against both strains of P falciparum while being inactive against human HepG2 cells. Also, the compound is predicted negative for three of the four toxicity models included. However, it triggered a CPDB Signature alert for carcinogenicity
This positive carcinogenicity prediction is not confirmed in the DS perspective with any of the OpenTox ‘IST DSSTox’ carcinogenicity or mutagenicity models, or the ‘ToxTree: Benigni/Bossa rules for carcinogenicity and mutagenicity’ model. The DS view offers additional information, though: the ‘ToxTree: Structure Alerts for the in vivo micronucleus assay in rodents’ is positive, and two ‘ToxTree: Skin sensitization alerts’ are triggered. Combining these pieces of information, we might decide against selecting this compound as a drug development candidate, even though we may reject the initial carcinogenicity prediction.
In terms of its cytotoxic experimental data, TCMDC-134695 is also a promising candidate, as shown in Figure 2.13. In addition, again three out of four Bioclipse Decision Support models are negative. However, the compound has a weak association predicted with human adverse events related to the hepatobiliary tract.
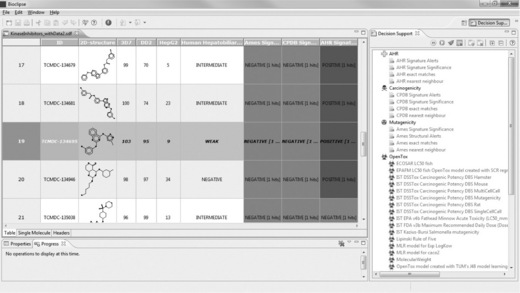
Figure 2.13 Molecule Table view shows TCMDC-134695 in row 19. This compound is promising for most of its properties represented in the table, except for the AHR Signature Significance. However, it is weakly associated with human adverse events
Running the OpenTox models in the DS perspective on this compound adds to the concerns raised with the predicted association with human adverse events. The compound is predicted to be carcinogenic and mutagenic with several OpenTox models (‘ToxTree: Structure alerts for the in vivo micronucleus assay in rodents’, ‘ToxTree: Benigni/Bossa rules for carcinogenicity and mutagenicity’, ‘IST DSSTox Carcinogenic Potency DBS Mouse’, ‘IST DSSTox Carcinogenic Potency DBS MultiCellCall’ and ‘IST DSSTox Carcinogenic Potency DBS Mutagenicity’).
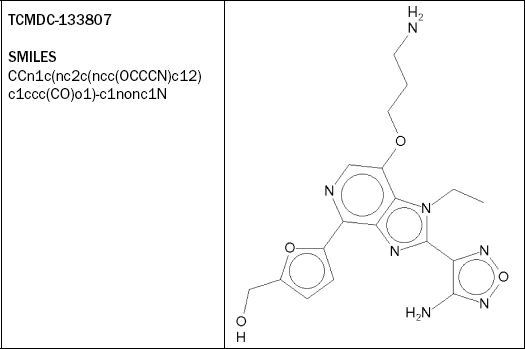
TCMDC-133807 (see Figure 2.14) appears to be a rather toxic compound, being predicted to be strongly associated with human adverse events and triggering a structure alert with Bioclipse’s CPDB carcinogenicity model.
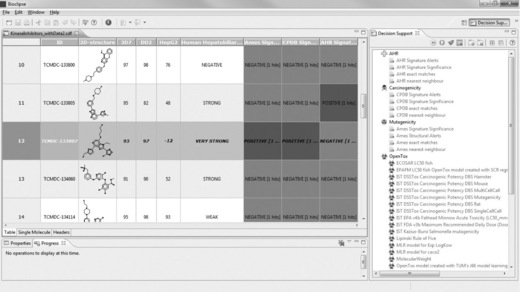
Figure 2.14 The compound TCMDC-133807 is predicted to be strongly associated with human adverse events, and yields signature alerts with Bioclipse’s CPDB and Ames models
Indeed the compound turns out positive with the following OpenTox models:
![]() ‘ToxTree: Structure alerts for the in vivo micronucleus assay in rodents’;
‘ToxTree: Structure alerts for the in vivo micronucleus assay in rodents’;
![]() ‘IST DSSTox Carcinogenic Potency DBS Mutagenicity’;
‘IST DSSTox Carcinogenic Potency DBS Mutagenicity’;
![]() ‘IST DSSTox Carcinogenic Potency DBS Rat’;
‘IST DSSTox Carcinogenic Potency DBS Rat’;
![]() ‘IST DSSTox Carcinogenic Potency DBS MultiCellCall’;
‘IST DSSTox Carcinogenic Potency DBS MultiCellCall’;
![]() ‘IST DSSTox Carcinogenic Potency DBS SingleCellCall’;
‘IST DSSTox Carcinogenic Potency DBS SingleCellCall’;
However, the compound is negative with the following OpenTox models:
![]() ‘IST DSSTox Carcinogenic Potency DBS Hamster’;
‘IST DSSTox Carcinogenic Potency DBS Hamster’;
![]() ‘IST DSSTox Carcinogenic Potency DBS Mouse’;
‘IST DSSTox Carcinogenic Potency DBS Mouse’;
![]() ‘IST DSSTox Carcinogenic Potency DBS Hamster’;
‘IST DSSTox Carcinogenic Potency DBS Hamster’;
![]() ‘ToxTree: Benigni/Bossa rules for carcinogenicity and mutagenicity’.
‘ToxTree: Benigni/Bossa rules for carcinogenicity and mutagenicity’.
The considerably higher number of positive than negative predictions confirms the initial toxicity estimation by the human adverse events model and Bioclipse’s Decision Support Models.
2.5 Discussion
These use cases show an interoperability advance, which enables toxicologists and people working in other life science fields to interactively explore and evaluate the toxicological properties of molecules. The integration into Bioclipse makes various features of the OpenTox platform available to the user, both via the GUI and the Bioclipse Scripting Language, the latter focusing on reproducibility, the former on interactive exploration and optimization. This dual nature of the Bioclipse-OpenTox interoperability makes it unique. A solution which is capable of dynamically discovering new services, applying them while working interactively, makes the platform different from other software like ToxTree [13], ToxPredict [32], and the OECD QSAR ToolBox [11], or more general tools like Taverna [33] and KNIME [34].
A further difference to some of these alternative tools is that the Bioclipse-OpenTox integration relies on semantic web technologies, which are seeing significant adoption in other areas of the life sciences too, including drug discovery, text mining, and neurosciences [35–37]. Several other chapters in this book describe applications making use of semantic web technologies. In particular David Wild’s work in creating the Chem2Bio2RDF framework (Chapter 18) and the chapter by Bildtsen and co-authors in the development of the ‘triple map’ application (Chapter 19). In addition, the chapters by Alquier (Chapter 16) and by Harland and co-authors (Chapter 17) describe application of semantic MediaWiki in life science knowledge management. The OpenTox platform has demonstrated the provision of a simple but well-defined and consistent ontology for the interaction with their services, providing functionality for both service discovery and service invocation. The SADI framework is the only known semantic alternative [38], but does not currently provide the same level of computational toxicology services as does OpenTox.
The combination of these two unique features makes it possible for Bioclipse–OpenTox to follow the evolution of computational toxicology closely and promptly. We are therefore delighted that this book chapter is available under a Creative Commons ShareAlike-Attribution license, allowing us to update it for changes in the Bioclipse-OpenTox environment.
2.6 Availability
Bioclipse with the Decision Support and OpenTox extensions can be freely downloaded for various platforms from http://www.bioclipse.net/opentox.
2.7 References
[1] European Parliament – European Commission, Regulation (EC) No. 1907/2006 of the European Parliament and of the Council. Official Journal of the European Union, 2006.
[2] Knudsen, T.B., Houck, K.A., Sipes, N.S., et al. Activity profiles of 309 ToxCast™ chemicals evaluated across 292 biochemical targets. Toxicology. 2011; 282(1–2):1–15.
[3] Schmidt, U., Struck, S., Gruening, B., et al, SuperToxic: a comprehensive database of toxic compounds. Nucleic Acids Research; 37. 2009:D295–D299. [Database issue].
[4] Kuhn, M., Campillos, M., Letunic, I., Jensen, L.J., Bork, P. A side effect resource to capture phenotypic effects of drugs. Molecular Systems Biology. 2010; 6:343.
[5] Williams-DeVane, C.R., Wolf, M.A., Richard, A.M. DSSTox chemical-index files for exposure-related experiments in ArrayExpress and Gene Expression Omnibus: enabling toxico-chemogenomics data linkages. Bioinformatics (Oxford, England). 2009; 25(5):692–694.
[6] Huang, D.W., Sherman, B.T., Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2009; 4(1):44–57.
[7] Kelder, T., Pico, A.R., Hanspers, K., et al. Mining biological pathways using WikiPathways web services. PloS One. 2009; 4(7):e6447.
[8] Kanehisa, M., Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Research. 2000; 28(1):27–30.
[9] Rydberg, P., Gloriam, D.E., Olsen, L. The SMARTCyp cytochrome P450 metabolism prediction server. Bioinformatics. 2010; 26(23):2988–2989.
[10] Willighagen, E.L., Jeliazkova, N., Hardy, B., Grafstrom, R.C., Spjuth, O. Computational toxicology using the OpenTox application programming interface and Bioclipse. BMC Research Notes. 4(487), 2011.
[11] Diderichs, R. Tools for Category Formation and Read-Across: Overview of the OECD (Q)SAR Application Toolbox. In: Cronin M.M.J., ed. In Silico Toxicology. RSC Publishing; 2010:385–407.
[12] Patlewicz, G., Jeliazkova, N., Safford, R.J., Worth, A.P., Aleksiev, B. An evaluation of the implementation of the Cramer classification scheme in the Toxtree software. SAR and QSAR in Environmental Research. 2008; 19(5–6):495–524.
[13] ToxTree, 2011. Available at http://toxtree.sourceforge.net
[14] Spjuth, O., Helmus, T., Willighagen, E.L., et al. Bioclipse: an open source workbench for chemo- and bioinformatics. BMC Bioinformatics. 2007; 8(1):59.
[15] Spjuth, O., Alvarsson, J., Berg, A., et al. Bioclipse 2: a scriptable integration platform for the life sciences. BMC Bioinformatics. 2009; 10(1):397.
[16] Spjuth, O., Eklund, M., Ahlberg Helgee, E., Boyer, S., Carlsson, L. Integrated decision support for assessing chemical liabilities. Journal of Chemical Information and Modeling. 2011; 51(8):1840–1847.
[17] Hardy, B., Douglas, N., Helma, C., et al. Collaborative Development of Predictive Toxicology Applications. Journal of Cheminformatics. 2010; 2(1):7.
[18] Jeliazkova, N., Jeliazkov, V. AMBIT RESTful web services: an implementation of the OpenTox application programming interface. Journal of Cheminformatics. 2011; 3:18.
[19] Spjuth, O., Alvarsson, J., Berg, A., et al. Bioclipse 2: a scriptable integration platform for the life sciences. BMC Bioinformatics. 2009; 10(1):397.
[20] Willighagen, E.L., Alvarsson, J., Andersson, A., et al. Linking the Resource Description Framework to cheminformatics and proteochemometrics. Journal of Biomedical Semantics. 2011; 2(Suppl 1):S6.
[21] Spjuth, O., Willighagen, E.L., Guha, R., Eklund, M., Wikberg, J.E.S. Towards interoperable and reproducible QSAR analyses: Exchange of datasets. Journal of Cheminformatics. 2(1), 2010.
[22] Steinbeck, C., Hoppe, C., Kuhn, S., et al. Recent developments of the chemistry development kit (CDK) – an open-source java library for chemo- and bioinformatics. Current Pharmaceutical Design. 2006; 12(17):2111–2120.
[23] Anon. CC0 1.0 Universal Public Domain Dedication. Available at: http://creativecommons.org/publicdomain/zero/1.0/,
[24] Goble, C.A., Bhagat, J., Aleksejevs, S., et al. myExperiment: a repository and social network for the sharing of bioinformatics workflows. Nucleic Acids Research. 2010; 38(suppl 2):W677–W682.
[25] Gamo, F.-J., Sanz, L.M., Vidal, J., et al. Thousands of chemical starting points for antimalarial lead identification. Nature. 2010; 465(7296):305–310.
[26] Faulon, J.-L., Visco, D.P.J., Pophale, R.S. The signature molecular descriptor. 1. using extended valence sequences in QSAR and QSPR studies. Journal of Chemical Information and Computer Sciences. 2003; 43(3):707–720.
[27] Carlsson, L., Helgee, E.A., Boyer, S. Interpretation of nonlinear QSAR models applied to Ames mutagenicity data. Journal of Chemical Information and Modeling. 2009; 49(11):2551–2558.
[28] Helgee, E.A., Carlsson, L., Boyer, S. A method for automated molecular optimization applied to Ames mutagenicity data. Journal of Chemical Information and Modeling. 2009; 49(11):2559–2563.
[29] Gellibert, F., Fouchet, M.-H., Nguyen, V.-L., et al. Design of novel quinazoline derivatives and related analogues as potent and selective ALK5 inhibitors. Bioorganic & Medicinal Chemistry Letters. 2009; 19(8):2277–2281.
[30] Adverse Event Reporting System (AERS). Available at: http://www.fda.gov/Drugs/GuidanceComplianceRegulatoryInformation/Suveillance/AdverseDrugEffects/default.htm.
[31] Pharmatrope, 2011. Available at http:/pharmatrope.com/
[32] OpenTox ToxPredict application, 2011. Available at http://toxpredict.org
[33] Oinn, T., Addis, M., Ferris, J., et al. Taverna: a tool for the composition and enactment of bioinformatics workflows. Bioinformatics. 2004; 20(17):3045–3054.
[34] Berthold, M.R., Cebron, N., Dill, F., et al. KNIME: The konstanz information miner. In: Preisach C., Burkhardt H., Schmidt-Thieme L., eds. Decker R eds. Data Analysis: Machine Learning and Applications. Springer; 2008:319–326.
[35] Ruttenberg, A., Clark, T., Bug, W., et al. Advancing translational research with the Semantic Web. BMC Bioinformatics. 8(Suppl 3), 2007.
[36] Splendiani, A., Burger, A., Paschke, A., Romano, P., Marshall, M.S. Biomedical semantics in the Semantic Web. Journal of Biomedical Semantics. 2011; 2(Suppl 1):S1.
[37] Willighagen, E.L., Brändle, M.P. Resource description framework technologies in chemistry. Journal of Cheminformatics. 2011; 3(1):15.
[38] Chepelev, L.L., Dumontier, M. Semantic Web integration of Cheminformatics resources with the SADI framework. Journal of Cheminformatics. 2011; 3:16.