6.7. A Random Coefficient Model
Another approach to the modeling of repeated measures data is to use an appropriate random coefficient model. In this approach, one or more regression parameters are assumed to be a random sample from a population of regression coefficients. These models are useful whenever the regression model for fitting the repeated data on a subject can be assumed to be a random variation of a population regression model. This approach can be especially useful due to its capacity to handle unequally spaced and/or unbalanced growth curve type data.
Let yiu be the piu×1 vector of repeated measures on the uth subject of the ith treatment group. Then consider a mixed effects model described as
where Xiu and Ziu are the known matrices of orders piu by q and piu by r respectively, and β is the fixed q by 1 vector of unknown nonrandom regression coefficients. The r by 1 vectors νiu are random effects with E(νiu)=0, and D(νiu)=σ2G1. Also ϵiu are the piu by 1 vectors of random errors with Eϵiu=0, Dϵiu=σ2Riu. The usual further assumption that the various variables are uncorrelated is also made.
When both Xiu and Ziu correspond to quantitative variables and Ziu is a submatrix of Xi, the model in Equation 6.17 is referred as a random coefficient model. The fact that Ziu is a submatrix of Xiu distinguishes this situation from the general mixed model (Equation 6.1) where no such assumption need be made. A situation where Ziu will be a submatrix of Xiu can be described formally as follows. Suppose for a certain random regression coefficient, say γiul, E(γiul)=βl, so that it can be written as
where νiul is random with E(νiul)=0. Thus any random regression coefficient νiu with E(ϵiu)=0 has its fixed effects counterpart, namely βl. Therefore, when the model is expressed in the matrix form as in Equation 6.17, the columns corresponding to βl and νiu in the matrices Xiu and Ziu respectively are identical, thereby making Ziu a submatrix of Xiu.
The random coefficient models provide ample flexibility to deal with the repeated measures data. Within-subject variability is conveniently dealt with by modeling it through the random errors ϵiu and the random slope coefficients νiu for changes in repeated measures specific to the uth subject in the ith treatment group. The correlation structures such as compound symmetry and autoregressive or unstructured covariances can be assumed for G1 and Riu, u=1,...,ni, i = 1,...,k. The development of an appropriate model and corresponding analysis can best be illustrated through an example.
EXAMPLE 8
Random Coefficients, A Pharmaceutical Stability Study This example is adopted from SAS/STAT Software: Changes and Enhancements through Release 6.12, pp. 684–685. The pharmaceutical stability data (used with permission from Glaxo Wellcome Inc.) presents replicate assay results as the observed responses for the shelf life of various drugs (in months). The response variable is potency of the drug relative to the percentage claim on the label. There are three batches of products which may differ in initial potency represented by intercepts and in degradation rates represented by the slope parameters. Since the batches are taken randomly, these intercepts and slope parameters are assumed to be the random coefficients. Note that piu, the number of repeated measurements on each subject, are not all equal. The model can be expressed as
where γ0i and γ1i respectively are random intercepts and slopes normally distributed with mean β0 and β1 respectively. We write γ0i=β0+νoi and γ1i=β1+νli. Then νoi and νli are normally distributed with zero means. Thus the above model can be reexpressed as
where ϵiju ~ N(0,σ2) are independent. The coefficients (νoi, νli)′ are assumed to be independently jointly distributed as bivariate normal with zero mean vector and variance covariance matrix σ2G1. The independence of (νoi,νli)′ and ϵi′ju is also assumed for all i, i′, j, u.
Let β=(β0, β1)′ and νi. For our example, we can provide an interpretation of the above model as follows. Since there are two random coefficients, namely the batch intercept and the batch slope in vector νi both of which may not have zero means, we write their effects as β+ν1, β+ν2.... In the interpretation, β is the fixed effect part, representing the mean initial potency and mean degradation rate, and the vectors ν1,ν2... are all 2 by 1 vectors with their variance-covariance matrix σ2G1, where G1 a 2 by 2 matrix, which we assume to be unstructured. The three unknown parameters of G1, namely g11, g12=g21, and g22, need to be estimated, along with several other parameters. For the error vector ϵiu= (ϵilu,....ϵipiuu)′ we assume the spherical covariance structure. We will use the restricted maximum likelihood (REML) procedure for the estimation of parameters of the variance covariance matrix.
The subject effect is represented by three batches and on each batch, data are collected at 0, 1, 3, 6, 9, and 12 months. As mentioned earlier, the age of the drug in months represents a fixed effect factor as well, along with an intercept in the model which can be interpreted as the mean initial potency.
To analyze this data using PROC MIXED, we essentially need to spell out these facts in compact form within short SAS code. Specifically, we must indicate that BATCH plays the role of SUBJECT (SUBJECT=BATCH); that INTERCEPT and AGE are random effects (RANDOM=INT AGE); and we specify only the fixed effects part of the model, which includes an intercept, (which need not be specified as SAS adds the intercept by default) and the variable AGE. The SAS code is given as Program 6.9 and the output is presented as Output 6.9.
/* Program 6.9 */
options ls = 64 ps = 45 nodate nonumber;
title1 'Output 6.9';
title2 'A Pharmaceutical Stability Study';
data rc;
input batch age@;
do i = 1 to 6;
input y@;
output;
end;
cards;
1 0 101.2 103.3 103.3 102.1 104.4 102.4
1 1 98.8 99.4 99.7 99.5 . .
1 3 98.4 99.0 97.3 99.8 . .
1 6 101.5 100.2 101.7 102.7 . .
1 9 96.3 97.2 97.2 96.3 . .
1 12 97.3 97.9 96.8 97.7 97.7 96.7
2 0 102.6 102.7 102.4 102.1 102.9 102.6
2 1 99.1 99.0 99.9 100.6 . .
2 3 105.7 103.3 103.4 104.0 . .
2 6 101.3 101.5 100.9 101.4 . .
2 9 94.1 96.5 97.2 95.6 . .
2 12 93.1 92.8 95.4 92.5 92.2 93.03 0 105.1 103.9 106.1 104.1 103.7 104.6
3 1 102.2 102.0 100.8 99.8 . .
3 3 101.2 101.8 100.8 102.6 . .
3 6 101.1 102.0 100.1 100.2 . .
3 9 100.9 99.5 102.5 100.8 . .
3 12 97.8 98.3 96.9 98.4 96.9 96.5
;
/*Source: Obenchain (1990). Data Courtesy of R. L. Obenchain*/
proc mixed data=rc;
class batch;
model y=age/s;
random int age/type=un sub=batch s;
run;Example 6.9. Output 6.9
A Pharmaceutical Stability Study
Covariance Parameter Estimates (REML)
Cov Parm Subject Estimate
UN(1,1) BATCH 0.97292750
UN(2,1) BATCH −0.10192674
UN(2,2) BATCH 0.03649300
Residual 3.30229533
Solution for Fixed Effects
Effect Estimate Std Error DF t Pr > |t|
INTERCEPT 102.70159884 0.64480457 2 159.28 0.0001
AGE −0.52417636 0.11845227 2 −4.43 0.0475
Solution for Random Effects
Effect BATCH Estimate SE Pred DF t
INTERCEPT 1 −0.99744294 0.68336297 78 −1.46
AGE 1 0.12668799 0.12362914 78 1.02
INTERCEPT 2 0.38582987 0.68336297 78 0.56
AGE 2 −0.20397070 0.12362914 78 −1.65
INTERCEPT 3 0.61161307 0.68336297 78 0.90
AGE 3 0.07728271 0.12362914 78 0.63
Solution for Random Effects
Pr > |t|
0.1484
0.3087
0.5740
0.1030
0.3735
0.5337Tests of Fixed Effects
Source NDF DDF Type III F Pr > F
AGE 1 2 19.58 0.0475 |
The output shows that the REML estimate of the matrix G1 is
and therefore, the REML estimator of G is
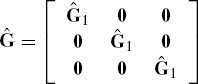
which is a 6 by 6 block diagonal matrix. Since the covariance structure for error is assumed to be spherical (σ2I), we do not need to specify this default choice in the SAS code. A REPEATED statement would be needed if any other covariance structure for error were to be specified. Under assumed sphericity, the estimated error variance is ![]() =3.3023.
=3.3023.
The effects of the intercept and slope are in part fixed and in part random. These are represented as ![]() , where β0 and β1 are the fixed parameters and ν0 and ν1 are the random coefficients, in each case respectively for the INTERCEPT and slope for AGE. The estimate of β and the predicted values of the random effects ν are presented in two separate tables in Output 6.9. Specifically,
, where β0 and β1 are the fixed parameters and ν0 and ν1 are the random coefficients, in each case respectively for the INTERCEPT and slope for AGE. The estimate of β and the predicted values of the random effects ν are presented in two separate tables in Output 6.9. Specifically,

Also presented are the corresponding standard errors, the prediction errors, and corresponding tests for significance. It may be remarked that since νi′s are random rather than fixed parameters, hypothesis testing on them may not be meaningful.
EXAMPLE 9
Modeling Linear Growth, Ramus Heights Data To further illustrate the use of the random coefficients models, we will analyze the ramus heights data of Elston and Grizzle (1962) where the heights of the ramus bone (in mm) for 20 boys were measured at 8, 8![]() , 9, and 9
, 9, and 9![]() years of age. We may want to model the ramus height, say yt, at age t as a polynomial growth function of their ages. Since these boys are a sample from a hypothetical population of all boys, the modeled growth curve can be thought of as the common growth curve for the population. However, due to many genetic and environmental variations, each boy would have his own individual growth curve which can be thought of as a random variation of the population growth curve.
years of age. We may want to model the ramus height, say yt, at age t as a polynomial growth function of their ages. Since these boys are a sample from a hypothetical population of all boys, the modeled growth curve can be thought of as the common growth curve for the population. However, due to many genetic and environmental variations, each boy would have his own individual growth curve which can be thought of as a random variation of the population growth curve.
For a given boy, we consider the model
where ϵt~ N(0,σ2) are independent and the values of β0 and β1 are specific to the specific boy. In other words, β0 and β1 are random coefficients. It is possible that only one of the two coefficients may be random. For example, if β0 is fixed and β1 is random then all the boys will have the common intercept of the population but different rate of growth. The difference is modeled as β1=β1F +β1R, where β1F is the fixed common slope for the population and β1R is the random part representing the amount of change from the common slope for the specific individual (boy). We assume E(β1R)=0 and var(β1R)=σβ12. Similarly, if β1 is fixed then all the boys will have the common rate of growth of the population but will have different intercepts. The appropriate assumptions to accommodate this case are β0=β0F+β0R, where β0F is the fixed common intercept for the population and β0R is the random part representing the amount of change from the common intercept for the specific individual (boy). We assume E(β0R)=0 and var(β0R)=σβ02. If both β0 and β1 are random then the above assumptions and interpretations hold for both coefficients. We will illustrate the case when only β1 is random.
The SAS code is presented as Program 6.10. For the sake of completeness we have also added the code for the other two cases in the same program (but they have been commented out).
If only β1 is random but β0 is fixed then the model (6.18) results in
Thus β0+β1F
aget represents the fixed part of the model and β1R is a normally distributed random coefficient with zero mean, and variance σβ12. Thus, σ2G1 is a 1 by 1 matrix, namely (σβ12). The errors ϵt are assumed to be independent and have a common variance, say, σ2. That is, the spherical structure for the variance covariance matrix of the errors is assumed. Since ![]() , or equivalently,
, or equivalently, ![]() , the option TYPE=SIM for the covariance structure of G1 can be used (in fact, this is a default option). Thus the appropriate MODEL and RANDOM statements are given by
, the option TYPE=SIM for the covariance structure of G1 can be used (in fact, this is a default option). Thus the appropriate MODEL and RANDOM statements are given by
model y=age/s; random age/type=sim subject=boy;
where the option S in the MODEL statement requests that the solution for the fixed effects be printed. The option SUBJECT = BOY indicates that the observations on a given BOY constitute the vector of repeated measures. We have chosen to use the METHOD=REML option as the choice of estimation procedure. The complete code is given in Program 6.10. The corresponding output appears as Output 6.10.
/* Program 6.10 */
options ls = 64 ps=45 nodate nonumber;
title1 'Output 6.10';
title2 'Analysis of Ramus Data';
data ramus;
input boy y1 y2 y3 y4;
y=y1;
age=8;
output;
y=y2;
age=8.5;
output;
y=y3;
age=9;
output;
y=y4;
age=9.5;
output;
lines;
1 47.8 48.8 49. 49.7
2 46.4 47.3 47.7 48.4
3 46.3 46.8 47.8 48.5
4 45.1 45.3 46.1 47.2
5 47.6 48.5 48.9 49.3
6 52.5 53.2 53.3 53.7
7 51.2 53. 54.3 54.58 49.8 50. 50.3 52.7
9 48.1 50.8 52.3 54.4
10 45. 47. 47.3 48.3
11 51.2 51.4 51.6 51.9
12 48.5 49.2 53. 55.5
13 52.1 52.8 53.7 55.
14 48.2 48.9 49.3 49.8
15 49.6 50.4 51.2 51.8
16 50.7 51.7 52.7 53.3
17 47.2 47.7 48.4 49.5
18 53.3 54.6 55.1 55.3
19 46.2 47.5 48.1 48.4
20 46.3 47.6 51.0 51.8
;
/* Source: Elston and Grizzle (1962). Reproduced by
permission of the International Biometric Society. */
proc mixed data=ramus covtest;
class boy;
model y=age/s;
random age /type = simple subject = boy;
title3 'Only slope is random';
run;
/*
proc mixed data=ramus covtest;
class boy;
model y=age/s;
random int /type = simple subject = boy;
title3 'Only intercept is random';
run;
proc mixed data=ramus covtest;
class boy;
model y=age/s;
random int age /type = simple subject = boy;
title3 'Both intercept and slope are random';
run;
*/Example 6.10. Output 6.10
Analysis of Ramus Data
Only slope is random
Covariance Parameter Estimates (REML)
Cov Parm Subject Estimate Std Error Z Pr > |Z|
AGE BOY 0.07944986 0.02647746 3.00 0.0027
Residual 0.66113890 0.12172554 5.43 0.0001
Solution for Fixed Effects
Effect Estimate Std Error DF t Pr > |t|
INTERCEPT 33.77000000 1.42583383 59 23.68 0.0001
AGE 1.86300000 0.17440771 19 10.68 0.0001Tests of Fixed Effects
Source NDF DDF Type III F Pr > F
AGE 1 19 114.10 0.0001 |
From Output 6.10 we find the estimates of variance components as ![]() =0.6611,
=0.6611, ![]() =0.0794, and
=0.0794, and ![]() =
= ![]() = 0.0525. It is appropriate to test the null hypothesis H0:σβ12=0 against the one-sided alternative. Testing H0:σβ12=0 is equivalent to testing H0: δ=0. Wald's test given in the output tests this null hypothesis against the two-sided alternative. The p value under the one-sided alternative can be computed as one-half of the reported p value. For our case, it is 0.0027/2≈ 0.0014, which is statistically significant. Thus we can claim σβ12 to be nonzero. Since E(yt)=β0+β1Faget, an estimate of average ramus height at time t is given by
= 0.0525. It is appropriate to test the null hypothesis H0:σβ12=0 against the one-sided alternative. Testing H0:σβ12=0 is equivalent to testing H0: δ=0. Wald's test given in the output tests this null hypothesis against the two-sided alternative. The p value under the one-sided alternative can be computed as one-half of the reported p value. For our case, it is 0.0027/2≈ 0.0014, which is statistically significant. Thus we can claim σβ12 to be nonzero. Since E(yt)=β0+β1Faget, an estimate of average ramus height at time t is given by
Suppose instead of fitting Equation 6.9, we decide to fit Equation 6.18 with a 4 by 1 vector of ϵt for each subject having a compound symmetric (CS) structure. In this case, there are no random effects in the model given in Equation 6.18, but the covariance structure within each subject would need to be specified using the REPEATED statement
repeated/type=cs subject=boy r;
where SUBJECT=BOY indicates that repeated measures which are assumed to be independent are taken on boys. Option R in the REPEATED statement prints a typical diagonal block of the block diagonal matrix R. However, we will not discuss this model further here.
