23
Medical Image Segmentation: Energy Minimization and Deformable Models
Chris McIntosh and Ghassan Hamarneh
CONTENTS
23.1 Definitions and Foundations
23.1.2 Segmentation Representation
23.1.6.1 Relation to Bayesian Methods
23.1.6.2 Relation to Segmentation via Registration
23.2 MIS via Energy Minimization
Appendix 23.B: Snakes: Details and Derivations
This chapter surveys the field of energy minimization as it applies to medical image segmentation (MIS). MIS remains a daunting task but one whose solution will allow for the automatic extraction of important structures, organs, and diagnostic features from medical images, with applications to computer-aided diagnosis, statistical shape analysis, and medical image visualization. Several classifications of segmentation techniques exist, including edge-, pixel-, and region-based techniques, in addition to clustering, and graph-theoretic approaches (Pham et al., 2000; Robb, 2000; Sonka and Fitzpatrick, 2000; Yoo, 2004). However, the unreliability of traditional, purely pixel-based methods in the face of shape variation and noise has caused recent trends (Pham et al., 2000) to focus on incorporating prior knowledge about the location, intensity, and shape of the target anatomy (Hamarneh et al., 2001). One type of approach that has been of particular interest to meeting these requirements is that of energy minimization methods due to their inherent ability to allow multiple competing goals to be considered.
In energy minimization methods, a function evaluates the goodness of a segmentation for a particular image, and the minimization of that function yields the segmentation of the image (Figure 23.1). Though highly variant in nature, the application of energy minimization methods to MIS is commonly built on five primary building blocks: (i) the energy function, (ii) the segmentation representation, (iii) the image representation, (iv) the training data, (v) and the minimizer. In what follows we provide an overview of each building block, and the major works therein over the past few decades.*
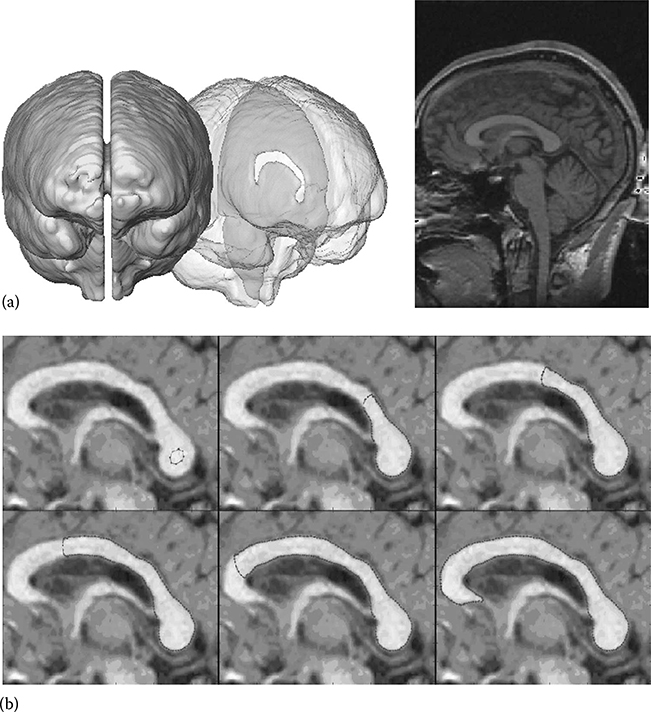
FIGURE 23.1
A corpus callosum (CC). (a) The CC is the band of nerve fiber tissue connecting the left and right hemispheres of the brain. (b) An energy minimization segmentation process. A shape model with progressively lower energy (left to right), showing a minimum of the energy function in bottom right. (Adapted from McIntosh, C. and Hamarneh, G., Vessel crawlers: 3D physically-based deformable organisms for vasculature segmentation and analysis, IEEE Computer Society Conference on Computer Vision and Pattern Recognition, 2006, Vol. 1, pp. 1084–1091, 2006a; Hamarneh, G. and McIntosh, C., Physically and statistically based deformable models for medical image analysis (Chapter 11), Deformable Models: Biomedical and Clinical Applications, pp. 335–386, 2007.)
23.1 Definitions and Foundations
We first give an outline of the energy minimization for MIS process. We define a medical image Ii and its corresponding segmentation (i.e., pixel labels) Si, each having N pixels. Then I = {I1, I2,…, Iℕ} and S = {S1, S2,…, Sℕ} are sets of images and their corresponding ground-truth segmentations. In a slight abuse of the notation, we occasionally omit the subscript i from Ii and Si for clarity and instead use I and S.
The first step in any energy minimization problem is the identification of the form of the energy function. In the next section, we will briefly group some popular energy terms into three main categories: boundary, region, and shape. Boundary terms are concerned primarily with the object boundary, region terms with the region inside or outside the object, and shape terms with the shape of the object. Other energy terms include spatial constraints on multipart objects, for example, containment (or layering), exclusion, or the number of labels (Delong et al., 2012; Nosrati and Hamarneh, 2013; Ulen et al., 2013; Yazdanpanah et al., 2011). Here we use these groupings to build a general energy functional. It may be convex or non-convex, as can the shape space over which it will be mini-mized. A general form is E(S, I, w) = w1 × boundary(S, I) + w2 × region(S, I) + w3 × shape Prior(S), with free parameter w = [w1, w2, w3]. Depending on the value of w, minima of E tend toward best satisfying the boundary, region, or shape terms. We note that the boundary and region terms are often referred to as external terms, since they involve cues external to the shape model, while the shape prior is deemed an internal term. More generally, we can write
where
Ji is the ith energy term
w = [w1,…,wn] are the weights
We note here that depending on the nature of S, E may be called an energy function or an energy functional, with the latter indicating S itself is a function.
The segmentation problem is to solve
which involves choosing a w and, depending on the nature of the energy functional, may also require training appearance and/or shape priors using I and/or S and setting an initialization. A gradient-descent-based solver is typically used but combinatorial approaches have also been explored for dis-cretized versions of the problem (Boykov and Kolmogorov, 2003) (see Section 23.5 for details). In either case, non-convexity, or supermodularity for combinatorial problems, can be quite problematic. There is no guarantee that another solution does not exist that better minimizes the energy and thus is potentially a better segmentation (Figure 23.2). Ideally both functional and shape spaces are convex, guaranteeing globally optimal solutions. However, whether convex or not the ground-truth segmentation, Si, for image, Ii is not guaranteed to be an optima (local or global) of E(S|Ii, w). The goal, in general, is to build an E(S|Ii, w) such that S* is as close as possible to Si under some definition of closeness [e.g., (Dice, 1945; Jaccard, 1901; Tanimoto, 1957)].
One of the earliest developed, and perhaps most recognized, examples of energy minimization methods being applied to image segmentation is that of deformable models. Deformable models for MIS gained popularity since the 1987 introduction of snakes by Terzopoulos et al. (Kass et al., 1987; Terzopoulos, 1987). At a high level, energy-minimizing deformable models work by deforming a user provided initial shape to fit to a target structure in a medical image. Shape-changing deformations result from the minimization, with respect to the shape, of a cost function measuring how plausible is the shape model and how well it aligns with the boundaries of the target anatomy in the image. Since the shape model itself is most commonly represented by a function, the cost function is often termed an energy functional and its gradient is derived using methods from variational calculus. The shape deformations are therefore typically simulated by solving an initial value problem using gradient-descent optimization algorithms (Elsgolc, 1962, G2). One further development, “Deformable organisms,” uses artificial life modeling techniques to augment the energy-minimizing deformable models with models of cognition, behav-iors, and sensory input (Hamarneh et al., 2001, 2009).
We now begin our more in-depth discussion of the different components of the energy minimization for MIS process.
23.1.1 Energy Function
Since snakes paved the way, there have been numerous papers attempting to increase accuracy by contributing novel energy terms, each designed to address a particular class of images or alleviate a particular problem with the original terms. As there have been far too many proposals to survey them all, here we focus on the key terms, many of which themselves have spawned numerous new approaches to MIS or changed the way we think about the problem at hand. As much of the initial development focused on external energy terms, namely, on the boundary and region terms that deal with the image processing itself, we begin our discussion there.
One of the most fundamental problems noted with snakes relates to their boundary capture range. If placed near a strong edge in an image, the contour would quickly snap to the edge, but if initialized farther away the influence of the edge would not reach the contour. There are, in fact, numerous causes of this problem relating to not only the external energy terms (Caselles et al., 1997; Cohen, 1991; Xu and Prince, 1998) but also the way the segmentation was originally represented (an explicit contour—see Section 23.1.2 for details) and the minimization technique being used.
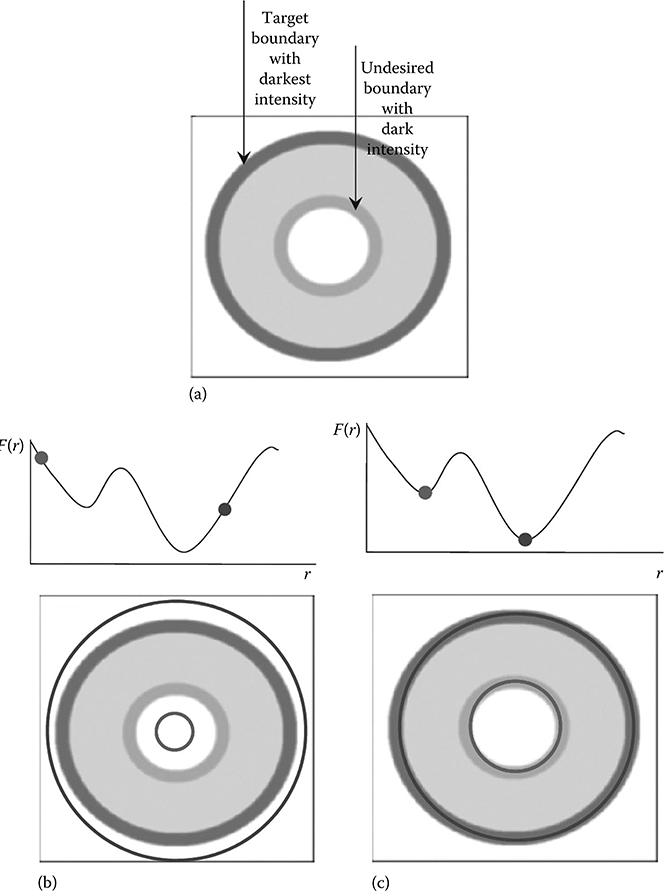
FIGURE 23.2
Synthetic example of single parameter deformable model with local minima. The circular deformable model's only parameter is its radius r. The energy function F(r) is minimal for the circle with darkest average intensity. The input image is shown in (a) with the darkest outmost boundary representing the global minima. In (b) two deformable models are initial-ized. In (c) after gradient descent, each model has moved to the nearest minima. (Reprinted from McIntosh, C., Energy functionals for medical image segmentation: Choices and consequences, PhD dissertation, Simon Fraser University, Burnaby, BC, Canada, Copyright 2011. With permission.)
An early attempt to rectify the aforementioned problem was by adding a deflation or inflation force to the contour that would attempt to shrink/grow it toward edges (Cohen, 1991). Rather than rely on a constant force, gradient vector flows extend the influence of edges off into homogeneous regions, thus increasing the capture range (Xu and Prince, 1998). Geodesic active contours (GACs) were similarly developed by both Caselles et al. (1997) and Yezzi et al. (1997). The approach of Caselles et al. formulated a deformable model optimization problem as that of finding the optimal path in a Riemannian space. Termed GACs, these popular deformable models work by minimizing curve length, where length is measured as the geodesic distance on a Riemannian manifold defined via an edge indicator function. The shortest curve is then, by definition, the curve along the edges of the image, and thus GACs automatically shrink the curve to the edges. GACs have become the canonical example of boundary-based deformable models.
However, what about objects whose boundaries blurred due to their inherent nature or noise, like the example in Figure 23.3? In these cases the intensity statistics of the areas both inside and outside the contour can be used to attempt to divide the image into maximally separated regions. The approaches of Chan et al. and of Yezzi et al. (Chan and Vese, 2001; Tsai et al., 2001) were similarly developed. Both are modeled after the Mumford–Shah functional wherein images are approximated by piecewise-smooth functions (Mumford and Shah, 1989). The approach of Chan and Vese is referred to as active contours without edges (ACWE) and has become a popular example of energy minimization for image segmentation. However, in their initial formulations, both methods approximate images by piecewise-constant functions, that is, objects are assumed to have a constant intensity. When objects are noisy, or their intensity changes gradually, a piecewise-smooth approximation is better suited, and thus an extension to piecewise-smooth functions was developed (Chan et al., 2007).

FIGURE 23.3
An energy minimization segmentation process. A shape model with progressively lower energy (left to right), showing a minima of the energy function in bottom right. Notice the leaking into neighboring structures that occurs as a result of weak edge strength. (Reprinted from McIntosh, C. and Hamarneh, G., 2006b, Genetic algorithm driven statistically deformed models for medical image segmentation, ACM Workshop on Medical Applications of Genetic and Evolutionary Computation Workshop (MedGEC), in Conjunction with the Genetic and Evolutionary Computation Conference (GECCO), Seattle, WA, 2006b. With permission.)
When both boundary- and region-derived statistics are not enough, shape-based terms are used. Shape terms provide resilience to false boundaries by heavily penalizing the implausible shape configurations that the false boundaries imply. The most basic terms attempt to achieve boundary smoothness by minimizing curve length, segmentation area, or curvature. More advanced terms compare the shape of the segmentation to some prior model of the shape in an effort to minimize their dissimilarity (Cootes et al., 1992, 1995, 2001; Cootes and Taylor, 1997; Cremers et al., 2001, 2002, 2008; Dambreville et al., 2006; Etyngier et al., 2007; Leventon et al., 2000; Paragios et al., 2006; Pohl et al., 2007; Vu and Manjunath, 2008; Warfield et al., 2000) (see Section 23.1.4 for discussions).
There has even been work trying to combine edge terms, piecewise-constant terms, and shape terms into a single formulation (Bresson et al., 2006). However, with so many competing assumptions about the object and the image inherent in each formulation, some level of trade-off must be established in the resulting functional (i.e., the weights, w, must be set).
If not appropriately set, the weights w can cause significant error. In fact, our results demonstrated that optimizing the weights has dramatic effects, reducing error in large data sets by as much as 60% (McIntosh and Hamarneh, 2007). However, optimizing the weights by hand for even a single image can be a long and tedious task, with no real guarantee of obtaining the correct segmentation. As such, there has been a number of works that seek to automatically set the weights (Anguelov et al., 2005; Finley and Joachims, 2008; Kolmogorov et al., 2007; McIntosh and Hamarneh, 2007, 2009; Szummer et al., 2008; Taskar et al., 2005 Rao et al., 2009, 2010).
Instead of guessing the optimal weights, suppose we write a function γ(w|Ij, Sj) evaluating how well weight w works for a given image-segmentation pair (Ij, Sj), such that a parameter is deemed better when it causes S* to approach Sj, that is, the minimum of E to be the correct segmentation. Given Sj, we could then calculate the ideal weights for a particular image Ij by solving . It is important that γ itself be convex or globally solvable in w. If γ was not globally solvable, uncertainty would remain in that another w* may better minimize γ and thus better segmentthe image. Similarly, γ cannot contain free parameters, else those parameters would themselves introduce uncertainty, as was the case in McIntosh and Hamarneh (2007).
Recent advances in maximum margin estimation allow for weight opti-mization wherein the parameters of the objective function are sought such that the highest scoring structures (in our case segmentations) are as close as possible to the ground truth (Anguelov et al., 2005; Finley and Joachims, 2008; Szummer et al., 2008; Taskar et al., 2005). These methods do however assume that a single set of weights works for the entire test set of images, whereas different images can easily require different weights. In contrast, Kolmogorov et al. seek an optimal parameter on a per-image basis. Given a parameter range, their method simultaneously solves the objective function for a set of parameters that bound how the parameters influence the solution (Kolmogorov et al., 2007). Each solution is then treated as a potential segmentation. They propose a number of heuristics, including user intervention, to select the best segmentation from a set of potential ones.
A related topic is that of multiobjective optimization, where methods try to jointly minimize the terms, without weighting them. As the topic relates more to minimizing energy functionals, we defer its discussion until Section 23.1.5.
23.1.2 Segmentation Representation
As expected, all functions must have domains and so in turn energy functionals must have domains. That domain is the space of possible segmentations of the image or shapes. There are many different ways to represent the underlying segmentation, and that choice in turn impacts the image and shape terms that can be readily evaluated. For example, a shape using closed contours pairs most readily with region statistics. Here we summarize some of the most prevalent shape representations in use.
Naturally, we start with the representation originally detailed by Terzopolous et al., namely the explicit contour model (Kass et al., 1987; Terzopoulos, 1987). The contour is defined as an explicit, parameterized function of its arc length. Integrating along the arc-length integrates along the contour and allows for the evaluation of both internal and external terms (see Appendix B for details). These methods were extended to represent not only contours but surfaces and volumes (Cohen et al., 1992; Cohen and Cohen, 1993; Delingette et al. 1992; McInerney and Terzopoulos, 1995a; Miller et al., 1991; Staib and Duncan, 1992b; Whitaker, 1994). Other explicit models were introduced relating to spring-mass systems, where each boundary point is a mass, connected to other masses via springs (McInerney and Terzopoulos, 1996; Montagnat et al., 2001). There can be other types of masses, namely, internal nodes and medial nodes (Pizer et al., 2003). However, in general these aforementioned works are constrained to a fixed topology, which posed problems in many applications.
There have been two major directions of work to address topological adaptability. The first direction focused on novel ways to automatically re-parameterize the contour or surface enabling the evolution into complex geometries (McInerney and Terzopoulos, 1995b,c, 1999, 2000). Initially developed in 2D, T-snakes work by subdividing the image domain into a Freudenthal triangulation (McInerney and Terzopoulos, 1995b,c). At each deformation step, the intersections of the contours with the triangular grids are found, and a set of rules is followed to determine if the contour point should be split or merged. This work was later extended to 3D, with the introduction of T-surfaces (McInerney and Terzopoulos, 1999, 2000). Delingette simultaneously developed a somewhat related approach where simplex meshes are used to model the shape, and topological changes are performed “semi-automatically with an automatic detection of topological problems, but a manual validation of all operations” (Delingette, 1999). However, in both cases the explicit re-parameterization of the contour or surface can be costly and may not generalize well to even higher dimensions.
In contrast, implicit contours were built from the ground up to handle changes in topology (Caselles et al., 1997; Osher and Paragios, 2003; Osher and Sethian, 1988; Sethian, 1996). In these representations, the boundary is implied by a given function, instead of explicitly defined. Implicit representations are built around the signed distance function (SDF), where the object boundary is defined as the zero level set of the function. Integrating over the domain of the SDF implicitly integrates over the contour and thus allows for the evaluation of both internal and external terms, as before. Changes in topology are handled internally by the shape representation and require no re-parameterization of the model (Figure 23.4). Similarly, other functions can be used to implicitly represent shapes including characteristic functions (Tsai et al., 2004) and probability maps (Cremers et al., 2008).
More recently, graph methods have emerged where the segmentation is represented by the assignment of labels to a graph (Barrett and Mortensen, 1997; Boykov and Funka-Lea, 2006; Boykov and Jolly, 2001; Boykov and Kolmogorov, 2003; Falcão et al., 1998; Falcão and Udupa, 1997; Grady, 2006; Mortensen and Barrett, 1998; Shi and Malik, 2000). Pixels are represented as vertices in the graphs, and edges between pixel–vertices represent a connectivity neighborhood. Energy functionals, often called cost functions in graph-based works, are expressed as sums over the vertices and their neigh-borhoods that vary as a function of how each vertex is labeled.
Finally, our discussion thus far has been limited to that of a single object class, but it is also possible to represent multiple object classes at one time, using so-called multi-class shape representations that are built on both implicit (Paragios and Deriche, 2000, 2002; Samson et al., 2000; Vese and Chan, 2001; Zhao et al., 1996) and graph-based representations (Boykov and Jolly, 2001; Grady and Funka-Lea, 2004). Implicit shape models are adapted to multiple classes by defining multiple implicit functions, where the combinations and intersections of the functions denote which class is being represented. Graph methods, however, are extended by increasing the number of possible labels for each vertex.

FIGURE 23.4
An exemplar SDF as a shape representation. (a) An SDF undergoing a simple deformation from left to right. (b) The zero level sets of the SDFs, displaying the segmentation each SDF represents. Notice how the topology automatically changes without any re-parameterization. (Reprinted from McIntosh, C., Energy functionals for medical image segmentation: Choices and consequences, PhD dissertation, Simon Fraser University, Burnaby, BC, Canada, Copyright 2011. With permission.)
In the end, the choice of representation is ultimately determined by the desired segmentation task. As such, it is worth noting that specialized models can exist, where a shape representation is designed specifically for one type of anatomy. A popular example is that of tubular-branching objects, namely, vessels, where cylindrical models can be built and deformed (McIntosh and Hamarneh, 2006; O'Donnell et al., 1994).
23.1.3 Image Representation
As might be expected, how the image is represented will impact how the terms of the energy functional can be evaluated on it. In fact, early on there were many contributions demonstrating how existing techniques could be modified to fit higher dimensional data, vector-valued data (Ballerini, 2001; Chan et al., 2000; Sapiro, 1996; Shah, 1996; Shi et al., 2008), tensor data (Nand et al., 2011; Rousson et al., 2004; Wang and Vemuri, 2004; Weldeselassie and Hamarneh, 2007) (Chapter 19), texture-heavy images (Paragios and Deriche, 2000, 2002), or even temporal data (Saad et al., 2008 Hamarneh et al., 2004; Tang et al., 2012; Rana et al., 2009; Ng et al., 2012), as opposed to the static, 2D, grayscale images early algorithms were presented on. Furthermore, numerous methods have been adapted to handle the intricacies of specific medical imaging modalities including magnetic resonance images and ultrasound. As an example, we talk briefly about some of the issues inherent to angiography in Section 23.2.
23.1.4 Training Data
A substantial amount of knowledge is often available about anatomical structures of interest—characteristic shape, position, orientation, symmetry, relationship to neighboring structures, associated landmarks, etc. and about plausible image intensity characteristics, subject to natural biological variability or the presence of pathology. Once collected, the training data typically come into the energy functional in the place of shape priors (Cootes et al., 1995), but appearance priors (Cootes et al., 2001) have also been developed (see Heimann and Meinzer (2009) for a complete survey). As shape priors have been a particular area of interest in the field, here we discuss some of the key works.
In many applications, prior knowledge about object shape variability is available or can be obtained by studying a training set of shape examples. This knowledge restricts the space of allowable deformations to a learned shape space that approximates the space of anatomically feasible shapes (Cootes and Taylor, 1997; Cootes et al., 1992, 1995, 2001; Cremers et al., 2001, 2002, 2008; Dambreville et al., 2006; Etyngier et al., 2007; Leventon et al., 2000; Paragios et al., 2006; Pohl et al., 2007; Vu and Manjunath, 2008; Warfield et al., 2000). One of the most notable works in this area is that of Cootes et al., where they introduced and refined active shape models (ASM) (Cootes et al., 1992, 1993, 1995). In ASM, principal component analysis (PCA) is calculated over a set of landmark points extracted from training shapes. The resulting principal components are used to construct a point distribution model (PDM) and an allowable shape domain (ASD). In a natural extension to their previous work, Cootes et al. modify their method to include image intensity statistics (Cootes et al., 2001). Staib and Duncan constrained the deformable models in Fourier space by conforming to probability distributions of the parameters of an elliptic Fourier decomposition of the boundary (Staib and Duncan, 1992a). Statistical prior shape knowledge was also incorporated in implicit, level set-based deformable models. Leventon et al. introduced statistical shape priors by using PCA to capture the main modes of variation of the level set representation (Leventon et al., 2000). However, as Pohl et al. point out, level sets do not form a vector space and hence more accurate shape statistics could be captured by transforming the shapes into a vector space using the logarithm of odds before performing PCA (Pohl et al., 2007).
Though simpler to optimize than their nonlinear counterparts, linear models of shape deformation may not always adequately represent the variance observed in real data. Linear shape models assume the data lies on a linear manifold, but shapes often lie on nonlinear manifolds where the manifold's properties are not accurately captured by linear statistics (Fletcher et al., 2004). For example, try fitting an ellipse to an “S”-like shape space. In order to include the entire letter, extraneous white space (nonvalid shapes) must also be included. Nonlinear shape models have been introduced to address this problem (Cootes and Taylor, 1997; Cremers, 2008; Cremers et al., 2001, 2002; Dambreville et al., 2006; Etyngier et al., 2007; Fletcher et al., 2004; Sozou et al., 1995).
However, we argue that the problem with linear statistics, as described earlier, is not necessarily due to the application of a linear model to nonlinear data but rather because of the implicit nature in which the statistics are applied (McIntosh and Hamarneh, 2011). By implicit we mean the statistics attempt to model variation in the shape, rather than variation in the parameters governing the deformations themselves. Note that we are not referring to the shape representation being implicit or explicit but instead whether the deformations are implicitly or explicitly studied. Implicit shape statistics result from the majority of previous deformable model approaches adopting a boundary-based shape representation, aside from a few exceptions (Pizer, 2003). As a consequence, the statistics are calculated using boundary models of the shape instead of models representing the interior and skeletal topology of the structures. Studying the underlying structural changes of a shape allows deformations that were previously non-convex to be decomposed into linear models. We refer to these as explicit shape statistics since they are calculated over the very parameters responsible for varying the object's shape. Consider an object represented by a single pixel. Different images of the object show the pixel moving around in a circle. A nonlinear function is required to describe the pixel's motion and hence no linear statistics can capture the motion adequately as long as it is the object's x, y position being studied. However, once decomposed into a function of sin and cos, the underlying parameter that controls the objects variability is linear in its variation, and hence linear statistics will yield greatly improved shape statistics. The same argument carries forward, albeit more complexly, to a more complex object. A simple bending of a shape's medial axis is a linear deformation under the appropriate representation, as it is simply a rotation of some of the medial nodes. However, the bending is a highly non-convex deformation once embedded in the image domain, as either an implicit shape (Pohl et al., 2007) or an explicit boundary-based model (Cootes et al., 1995).
23.1.5 Minimizer
Once the image and shape representations have been set and the problem formulated, all that is left is to solve the minimization process and obtain the resulting segmentation. Though it may sound simple enough, this area has been a major focus of criticism of energy minimization-based methods over the years, and as such has recently become one of the most focused areas for research. Specifically, in their original inception, many of the aforementioned methods were plagued by problems of local minima and sensitivity to initialization. Here we describe the changes and revelations in the field on this topic over the past decade. For a broader review, the interested reader is referred to the following representative, but far from comprehensive, list: Cremers et al., 2011; Grady and Polimeni, 2010; Kolmogorov and Zabin, 2004; and Nikolova et al., 2006.
Energy functional minimization can be carried out in a variety of ways. One solution is to perform explicit differentiation under the Euler–Lagrange equations, where each new application with a modified energy functional must be accompanied by one such derivation (Caselles et al., 1997; Chan and Vese, 2001; Kass et al., 1987; Terzopoulos, 1987) (see Appendix A for details). The result is a set of equations, which, if satisfied, guarantee a stationary point of the energy functional. The solution is then obtained through a gradient-descent process where the change in the shape model (with respect to an artificial time variable) is equated to the Euler–Lagrange equation, that is, the deformable models come to rest when the equations are satisfied (Kass et al., 1987; Terzopoulos, 1987). There are, however, two main drawbacks with this approach. Firstly, performing gradient descent in the presence of image noise can lead to instability in the deformations over time (Sundaramoorthi et al., 2007). Secondly, as the number of dependent variables (shape, location, scale, orientation, etc.) increases so does the complexity of the search space, which often increases the number of local min-ima and requires the calculation of an increasingly large number of derivatives.
On the issue of stability, there has been work by Sundaramoorthi et al. on reformulating the gradient flow using Sobolev-type inner products, which induce favorable regularity prosperities into the flow, thus bringing smoother deformations over time (Sundaramoorthi et al., 2007). Recently Bar and Sapiro introduced a Newton-type method built on more general-ized norms than the L2 norm, obtained by equating the Euler–Lagrange constraint to an artificial time variable (Bar and Sapiro, 2009). They include the Sobolev norm and demonstrate improved results over L2 norms, with faster convergence and more accuracy in the presence of noise. Unless used on convex problems, however, these methods are still prone to local minima.
If local minima do not suffice, global minima must be sought, and as such there have been a number of recent approaches to obtaining the global optima of energy functionals (Andrews et al., 2011b; Appleton and Talbot, 2006; Barrett and Mortensen, 1997; Boykov and Funka-Lea, 2006; Boykov and Kolmogorov, 2003; Bresson et al., 2007; Cremers et al., 2008; Falcão and Udupa, 1997; Falcão et al., 1998; Grady, 2006; Li and Yezzi, 2006; Mortensen and Barrett, 1998; Nikolova et al., 2006). There are three primary directions toward this goal: min-paths, min-cuts, and convex approximations.
Min-path techniques are formulated on the basis of Dijkstra's algorithm for finding the shortest path in an undirected graph with nonnegative edge weights. They were first presented for 2D interactive segmentation (Barrett and Mortensen, 1997; Falcão and Udupa, 1997), extended to 3D (Falcão and Udupa, 1997), and later specialized to 4D for vessel segmentation (Li and Yezzi, 2006; Poon et al., 2008, kawahara et al., 2013, Wink et al., 2004) and 6D (or higher) for spinal cord segmentation (Kawahara et al., 2013).
Graph cuts were demonstrated as a global minimization technique for a popular energy functional (Caselles et al., 1997), as a special case of computing a geodesic on a Riemannian space whose metric is computed from the image (Boykov and Kolmogorov, 2003). However, graph cuts have been shown to apply only to a restricted class of energy functionals that are submodular (Kolmogorov and Zabin, 2004), and their solutions are discrete approximations to the continuous formulations whose accuracy is dependent on the resolution of the approximating graph (Boykov and Kolmogorov, 2003). Naturally, as that resolution increases so does their running time. Random walkers were developed in a similar nature, solving image segmentation as a graph problem wherein the global optimum is obtained to a particular cost function (Grady, 2006). In fact, graph cuts and random walkers have been shown to be specific instantiations of a single framework (Sinop and Grady, 2007).
Another line of work has come from the relaxation of the underlying shape model from a non-convex space to a convex one, thereby defining convex energy functionals that can then be minimized instead of their non-convex counterparts. This convex relaxation work, which began in 2004 with a simple restricted class of functionals (Nikolova et al., 2006), was later extended to a broader class in Bresson et al. (2007), and then a similar work appeared in 2008 with the addition of a shape prior (Cremers et al., 2008). However, restrictions still exist in that the functionals and the shape spaces they are optimized over must be convex when defined over the relaxed space and that the relaxed shape space must itself be convex.
Though not guaranteed to find global optima, genetic algorithms (GAs) have also been applied to the minimization of energy functionals (Ballerini, 1998, 2001; Fan et al., 2002; Ibáñez et al., 2009; MacEachern and Manku, 1998; McIntosh and Hamarneh, 2011; Tohka, 2001). At a high level, GAs work by performing many simultaneous local searches, each individually optimizing the energy functional via a random walk in the search space. At the end of the process, the search that yielded the lowest value for the energy functional is adopted as the segmentation.
Ballerini extends the classical active contour models, developed by Terzopoulos (1987), by using GA to directly minimize the standard energy functional (Ballerini, 1998). Members of the GA population are hypothetical shape configurations, represented by their explicit contour locations. The method was later extended to color images by using one image term per color channel (Ballerini, 2001). MacEachern and Manku presented a similar method using a binary representation of the contour (MacEachern and Manku, 1998). Similarly, Tohka presented simplex meshes paired with image-based energies, minimized via a hybrid GA-greedy approach, and applied the technique to the segmentation of 3D medical images (Tohka, 2001). Fan et al. also developed a GA method for an explicit active contour but describe their method using Fourier descriptors and employ parallel GAs to speed up minimization (Fan et al., 2002). A different shape representation, known as topological active nets, is used by Ibáñez et al. to enable the segmentation of objects with unknown topologies or even multiple objects in the same scene (Ibáñez et al., 2009). However, aside from simple boundary smoothness constraints, all of these methods are based on classical active contour models or their variants without incorporating prior shape knowledge, making them prone to latching to erroneous edges and ill-equipped to handle gaps in object boundaries (as was discussed in Section 23.1.1).
In Hill and Taylor (1992), GAs were used with statistically based ASMs, where the parameter space consists of possible ranges of values for the pose and shape parameters of the model. The objective function to be maximized reflects the similarity between the gray levels related to the object in the search stage and those found from training. Additional works use convex, implicit, global shape statistics assuming a Gaussian distribution around a mean shape (Ghosh and Mitchell, 2006; Mignotte and Meunier, 1999; Ruff et al., 1999). Mignotte and Meunier (1999) incorporate prior shape information by defining the mean as a circular deformable template, while Ruff et al. (1999) use a PDM for occluded shape reconstruction, and Ghosh and Mitchell (2006) use a level set shape representation and a learned mean from training data. Although these techniques apply GAs to produce generations of plausible populations of shapes, the statistically based deformations are convex and may not offer the required flexibility to accommodate for nonlinear shape deformations. In McIntosh and Hamarneh (2011), we address this problem by using GAs to optimize statically based deformations that explicitly study the underlying shape variations, thus reducing the problem with linear shape statistics described in Section 23.1.4.
A somewhat related direction is that of multiobjective optimization, where each term is simultaneously optimized rather than optimizing a linear sum of the terms (23.1). Hence, in multiobjective optimization no weights are provided to combine the competing terms of the functional, instead a solution is sought for which no term can be improved without sacrificing another (Collette and Siarry, 2002). That solution is known as a Pareto optimal solution (Collette and Siarry, 2002). The set of Pareto optimal solutions for a given problem is known as the Pareto front, and there is no preference among them unless a ranking is provided between the objectives. Nakib et al. recently used multiobjective optimization to determine the parameters for a thresholding algorithm for image segmentation (Nakib et al., 2010). Hanning et al. present an approach using a piecewise approximation of the image similar to the Mumford–Shah model (Mumford and Shah, 1989) using multiobjective optimization to decide the trade-off between the number of segments and the image approximation error (Hanning et al., 2006). It is interesting to note that, for convex functions, if the ground truth lies on the Pareto front, then by definition a set of weights must exist that causes the optima to be the ground truth for the linear sum of terms formulation (23.1) (Geoffrion, 1968). In other words, a linear sum of terms model exists that yields the same segmentation as the multiobjective model, which is far more challenging to optimize. However, the necessary weights needed to achieve the desired segmentation are unknown. Weight optimization attempts to determine these weights (Section 23.1.1).
23.1.6 Related Methods
Though our discussion thus far has been focused on energy minimization methods for image segmentation that follow the prescribed building blocks, there are a few bodies of related research that follow a different path. Here we briefly detail two of them.
23.1.6.1 Relation to Bayesian Methods
Throughout this chapter, we have examined numerous methods built on energy functionals of the form E(S|I,w) = w1 × internal(S) + w2 × external(S,I). However, an interesting parallel to probabilistic approaches can be observed with a few simple assumptions and manipulations of this general form. Here we demonstrate that this model is actually equivalent to performing image segmentation via Bayesian inference. First, we restate the segmentation problem as
where
Maximizing (23.3) is equivalent to minimizing its negative logarithm
where = −log P(I|S, w) − log P(S|w), and the denominator has been removed as it has no consequence on the minimization. Now suppose we model our probabilities as
then substituting back into (23.5) yields
as before. From here it becomes possible to examine many of the approaches previously cited and see what independence assumptions they are making from a Bayesian standpoint.
23.1.6.2 Relation to Segmentation via Registration
Thus far we have discussed methods where the dependent function of the energy functional is one describing the shape of the current segmentation. However, a related field exists where the dependent function is instead a spatial transformation describing how one or more images are related. This is the field known as image registration, and it involves finding a mapping from the spatial coordinates of one image to another, identifying which pixels in a source image map to which pixels in a target image. Image registration can be used for segmentation when the mapping from a novel image is found to a training image with a known segmentation, since that mapping effectively labels the novel image. As this field is far too large and diverse to cover here, we refer the interested reader to Maintz and Viergever (1998), Zitová and Flusser (2003), and Chapter 22, for a complete survey. We do note, however, that many of the same issues discussed in this chapter, that is, global versus local optima and setting the weights for the energy functional, are also important problems in registration.
23.2 MIS via Energy Minimization
As already noted in this chapter, energy minimization methods have been applied to a wide variety of MIS problems. Two popular application domains are those of cardiac images and vascular images. As an example of how energy minimization can be applied to MIS, here we briefly discuss some key works relating to vascular segmentation. Though a complete survey of energy mini-mization for MIS does not exist, the interested reader is referred to McInerney and Terzopoulos (1996) for a related survey of MIS using deformable models.
One structure of particular interest in the diagnosis and understanding of many diseases is vasculature (Bullitt et al., 2003). Vessel segmentation remains an interesting application area of energy minimization methods because of its unique challenges. Firstly, the topology is complex, and as such many of the already mentioned topology-adaptive shape models were first demonstrated in their application to vessel segmentation (McInerney and Terzopoulos, 2000). Secondly, the vessels are often of very low contrast motivating advances in image terms (Frangi et al., 1999; Vasilevskiy and Siddiqi, 2002; Wink et al., 2001, 2004). For example, Vasilevskiy and Siddiqi built flux-maximizing geometric flows based on the observation that the gradient vector field near a vessel should be orthogonal to the vessel (Vasilevskiy and Siddiqi, 2002). They define a flux-maximizing geometric flow as one for which the inward normals of the underlying curve align with the direction of the gradient vector field. Near vessels the gradient vector field points inward toward the vessel centerline, and thus maximizing the flux will align the boundary of the segmentation to the boundary of the vessel. The last major challenge in vessel segmentation is that the vessels can be very thin, pushing the boundaries of numerical stability in many techniques and motivating new methods with increased stability to thin structures (Lorigo et al., 2001; Sundaramoorthi et al., 2007). For example, Lorigo et al. modify GAC to deform along the medial axis of a tubular shape, as opposed to its surface (Lorigo et al., 2001). For a complete survey of vessel segmentation techniques, the reader is referred to Lesage et al. (2009).
23.3 Discussion
Having briefly touched on each of the five primary building blocks of energy minimization methods, we conclude with a discussion of the main issues concerning their usage for MIS, namely, issues relating to how to build the energy functional, represent the segmentation, deal with the different imaging modalities themselves, train priors, and finally minimize the resulting system. We begin by touching briefly on issues relating to validating the method, as that is a fundamental step that we have not yet discussed.
Every MIS method must be validated. There are two main approaches to validate an MIS method, expert segmentations and synthetic data, each of which has its own inherit advantages and drawbacks. Expert segmentations can be time consuming and costly to obtain. Furthermore, expert segmentations suffer from both inter- and intraoperator variability because multiple experts, or even the same expert on different days, can obtain differing segmentations of the same object. Warfield et al. attempt to address problems with inter- and intraoperator variability through an expectation–maximization algorithm that weights different segmentations according to a variety of measurements and rules (Warfield et al., 2004). The key advantage to expert segmentations of real medical data is that the data is real and hence it demonstrates the applicability of the method to the problem at hand. Synthetic data can be created by either physical or computational phantoms. Physical phantoms are those physically constructed and then imaged in some manner, whereas computational phantoms are simulated using mathematical models designed to replicate human anatomy under specific imaging protocols (Cocosco et al., 1997; Hamarneh and Jassi, 2010). The main drawback in both cases is realism: segmenting a phantom well does not necessarily mean real data will also be segmented with high accuracy. The main advantage of synthetic data is certainty about the ground-truth segmentations. In Hamarneh et al. (2008), a hybrid method based on synthesizing deformation of a real data is presented. Whether real data or synthetic data, a measure of dissimilarity between the automatic segmentation and the ground truth must eventually be computed. Standard approaches for evaluating segmentation results given ground-truth segmentation include the Hausdorff distance, the Dice similarity coefficient (Dice, 1945), the Jaccard index (Jaccard, 1901), and the Tanimoto coefficient (Tanimoto, 1957).
Choosing the right energy functional for the given task is a crucial first step. Ideally, one hopes for strictly convex functions with their global minima lying at the correct segmentations for a given set of images, but this ideal scenario is rarely the case. The main challenges here are determining what energy terms could make a good functional and how to weight them in a such way as to best segment the images, In other words, one must appreciate the trade-off between the fidelity of the energy functional (how accurately it models the segmentation problems at hand) and its optimizability (how attainable is the functional's global optimum) (Hamarneh et al., 2011; Ulen et al., 2013).
There are often two main concerns when choosing the segmentation representation. Firstly, will it allow for the training of appropriate shape priors? Secondly, will it introduce problems in the minimization stage? Some shape representations do not form vector spaces, and thus performing statistics on them is difficult. Level sets (SDFs) are one such example, where a specific method for computing statistics over the resulting shape space was needed (Pohl et al., 2007). Level set-based representations can also cause problems in the minimization stage, as they do not form a convex space, and thus introduced non-convexity into the minimization problem (Cremers et al., 2008). However, they remain a popular technique due to the sub-pixel accuracy and automatic handling of topological changes. A more recent representation is the isometric log ratio (Changizi and Hamarneh, 2010) that has been used for segmentation of anatomical images (Andrews et al., 2011a,b).
Each imaging modality has its own inherent problem associated with it relating to different types of noise and spatial or temporal resolution of the structure of interest. As already exemplified with a brief treatment of blood vessel segmentation methods, each application can require customized energy functionals, segmentation representations, and optimization strategies.
When training priors, one is often forced to balance two competing goals: having the priors accurate versus having the resulting energy functional solvable. Linear statistics do not often fit the training data well but lead to easily solved energy functionals, whereas nonlinear statistics fit the data but are difficult to work with. The problem with linear statistics, however, is often not necessarily due to the application of a linear model to nonlinear data but rather due to the global aspect of the shape statistics themselves and due to the shape representation to which they are applied. Global shape statistics are those that model the shape variation of the entire shape simultaneously, that is, each shape is a single point in some high-dimensional space and the statistics (linear or not) describe some restricted set of that space. Many deformable model approaches adopt a boundary-based representation. As a consequence the statistics are calculated using boundary models of the shape instead of models representing the interior and skeletal topology of the structures, leading to a loss of accuracy (McIntosh and Hamarneh, 2011).
The primary problem in minimization remains how to globally optimize an increasingly large set of energy functionals, as opposed to the restricted sets seen thus far (Bresson et al., 2007; Cremers et al., 2008; Kolmogorov and Zabin, 2004). Interestingly enough, even with global minima, existing functionals are not proving accurate enough, motivating the search for new energy terms or ways of building functionals that can better respect the image variability.
In short, developing MIS methods remains a daunting task from start to finish with numerous areas for research relating to each of the five building blocks commonly found in energy minimization techniques. However, with the ever-growing popularity of medical imaging for diagnosis and disease understanding, developing robust and automatic techniques for MIS remains an important goal. In order to reach that goal, we must continue to make breakthroughs in terms of accuracy and reliability of MIS methods. To do that, we need to consider where the bulk of the error and performance variability is coming from, so as to best focus our efforts there.
With fully automatic segmentation remaining an unsolved problem, user-based inspection and delineation of medical images is indispensable. This is trivial for scalar fields, but for complex manifold-valued images, even displaying these images requires careful considerations.
Realizing the challenge in achieving accurate yet fully automatic segmentation methods, recent methods opted to calculate the uncertainty in a resulting segmentation (e.g., via Shannon's entropy of probabilistic segmentation) and utilizing the uncertainty measures in active- and self-learning strategies (Andrews et al., 2011a,b; Changizi and Hamarneh; 2010, Saad et al., 2010a,b; Top et al., 2010, 2011).
Appendix 23.A Euler–Lagrange
Starting with a function Φ(x), we build an energy functional
where Φx is the first derivative of Φ with respect to x and Φxx the second. The functional E describes a desirable feature of Φ, in that it takes a high value when Φ is doing poorly and a low value when Φ is doing well. We know from Fermat's theorem that E obtains its extremum at a stationary point, that is, where the derivative is zero. To describe a stationary point one typically writes
where ω is a scalar, but here ω is a function, so this equation is known as the first variation, as opposed to fist derivative, of the functional. Developed in the 1750s, the Euler–Lagrange equation plays a fundamental role in variational calculus, defining a necessary condition under which the first variation tends to zero, and therefore the functional reaches a stationary point (Elsgolc, 1962). For a functional E as described earlier, the general form of the Euler–Lagrange differential equation is
See Appendix B for an example of its application to energy minimization methods for MIS.
Appendix 23.B Snakes: Details and Derivations
Classical deformable shape models (Terzopoulos, 1987) are represented as a 2D parametric contour v(s) = (x(s), y(s)), where s ∊ [0,1] traverses the contour (Figure 23.B.1) and v is deformed to fit to image data by minimizing an energy term ξ
which depends on both the shape of the contour and the image data I(x, y) reflected via the internal and external energy terms, α(v(s)) and β(v(s)), respectively.
The internal energy term is given as

FIGURE 23.B.2 A parameterized contour undergoing a simple deformation (left to right). (Reprinted from McIntosh, C., Energy functionals for medical image segmentation: Choices and consequences, PhD dissertation, Simon Fraser University, Burnaby, BC, Canada, Copyright 2011. With permission.)
whereas the external energy term is given as
The weighting functions w1 and w2 control the tension and flexibility of the contour, respectively, and w3 controls the influence of image data. wi’s can depend on s but are typically set to different constants. For the contour to be attracted to image features, the function P(v(s)) is designed such that it has minima where the features have maxima. For example, for the contour to be attracted to high-intensity changes (high gradients), we can choose
where Gσ * I denotes the image convolved with a smoothing (e.g., Gaussian) filter with a parameter σ controlling the extent of the smoothing (e.g., variance of Gaussian).
The contour v that minimizes the energy ξ must, according to the calculus of variations (Elsgolc, 1962), satisfy the vector-valued partial differential (Euler–Lagrange) equation
where . The first step in applying the Euler–Lagrange is to determine the partial derivatives as follows:
Then substituting back into Equation 23.B.5, we get the final set of equations
where we make note that v(s) = (x(s), y(s)), and so the earlier equation can be expanded to
In order to solve the system of equations, we introduce an artificial time step, by equating the earlier equations to −∂v/∂t. This yields a first-order iterative optimization method, though as outlined in this report other choices for optimization methods exist.
References
Andrews S., Hamarneh G., Yazdanpanah A., HajGhanbari B., and Reid D., 2011a, Probabilistic multi-shape segmentation of knee extensor and flexor muscles, in Lecture Notes in Computer Science, Medical Image Computing and Computer-Assisted Intervention (MICCAI), Vol. 6893, pp. 651–658.
Andrews S., Hamarneh G., Yazdanpanah A., HajGhanbari B., and Reid W.D., 2011a, Probabilistic multi-shape segmentation of knee extensor and flexor muscles, Lecture Notes in Computer Science, Medical Image Computing and Computer-Assisted Intervention (MICCAI), pp. 651–658.
Andrews S., McIntosh C., and Hamarneh G., 2011b, Convex multi-region probabilistic segmentation with shape prior in the isometric log-ratio transformation space, IEEE International Conference on Computer Vision (IEEE ICCV), Barcelona, Spain.
Andrews S., McIntosh C., and Hamarneh G., 2011b, Convex multi-region probabilistic segmentation with shape prior in the isometric Logratio transformation space, in IEEE International Conference on Computer Vision (IEEE ICCV), pp. 2096–2103.
Anguelov D., Taskarf B., Chatalbashev V., Koller D., Gupta D., Heitz G., and Ng A., 2005, Discriminative learning of Markov random fields for segmentation of 3D scan data, IEEE Computer Society Conference on Computer Vision and Pattern Recognition, 2005, CVPR 2005, Vol. 2, pp. 169–176, San Diego, CA.
Appleton B. and Talbot H., 2006, Globally minimal surfaces by continuous maximal flows, IEEE Transactions on Pattern Analysis and Machine Intelligence 28, 106–118.
Ballerini L., 1998, Genetic snakes for medical image segmentation, Proceedings of the SPIE Conference on Mathematical Modeling and Estimation Techniques in Computer Vision, Vol. 3457, pp. 284–295, San Diego, CA.
Ballerini L., 2001, Genetic snakes for color images segmentation, in Applications of Evolutionary Computing (ed. Boers E.), Vol. 2037 of Lecture notes in Computer Science, Springer, Berlin/Heidelberg, Germany, pp. 268–277.
Bar L. and Sapiro G., 2009, Generalized Newton-type methods for energy formulations in image processing, SIAM Journal of Imaging Science 2(2), 508–531.
Barrett W.A. and Mortensen E.N., 1997, Interactive live-wire boundary extraction, Medical Image Analysis 1(4), 331–341.
Boykov Y. and Funka-Lea G., 2006, Graph cuts and efficient N-D image segmentation, International Journal of Computer Vision (IJCV) 70(2), 109–131.
Boykov Y. and Jolly M.P., 2001, Interactive graph cuts for optimal boundary & region segmentation of objects in n-d images, Proceedings of the Eighth IEEE International Conference on Computer Vision, 2001, ICCV 2001, Vol. 1, pp. 105–112, Vancouver, BC, Canada.
Boykov Y. and Kolmogorov V., 2003, Computing geodesics and minimal surfaces via graph cuts, Proceedings of the Ninth IEEE International Conference on Computer Vision, 2003, Vol. 1, pp. 26–33, Nice, France.
Bresson X., Esedoḡlu S., Vandergheynst P., Thiran J.P., and Osher S., 2007, Fast global minimization of the active contour/snake model, Journal of Mathematical Imaging and Vision 28(2), 151–167.
Bresson X., Vandergheynst P., and Thiran J.P., 2006, A variational model for object segmentation using boundary information and shape prior driven by the Mumford-Shah functional, International Journal of Computer Vision 68(2), 145–162.
Bullitt E., Gerig G., Aylward S., Joshi S., Smith K., Ewend M., and Lin W., 2003, Vascular attributes and malignant brain tumors, Medical Image Computing and Computer-Assisted Intervention—MICCAI 2003, Vol. 2878 of Lecture notes in Computer Science, Springer, Berlin/Heidelberg, Germany, pp. 671–679.
Caselles V., Kimmel R., and Sapiro G., 1997, Geodesic active contours, International Journal of Computer Vision 22, 61–79.
Changizi N. and Hamarneh G., 2010, Probabilistic multi-shape representation using an isometric log-ratio mapping, in Lecture Notes in Computer Science, Medical Image Computing and Computer-Assisted Intervention (MICCAI), Vol. 6363, pp. 563–570.
Changizi N. and Hamarneh G., 2010, Probabilistic multi-shape representation using an isometric log-ratio mapping, Lecture Notes in Computer Science, Medical Image Computing and Computer-Assisted Intervention (MICCAI), pp. Part III: 563–570.
Chan T., Moelich M., and Sandberg B., 2007, Some recent developments in variational image segmentation, Image Processing Based on Partial Differential Equations, Springer, Berlin, Germany, pp. 175–210.
Chan T. and Vese L., 2001, Active contours without edges, IEEE Transactions on Image Processing 10(2), 266–277.
Chan T.F., Sandberg B.Y., and Vese L.A., 2000, Active contours without edges for vector-valued images, Journal of Visual Communication and Image Representation 11(2), 130–141.
Cocosco C.A., Kollokian V., Kwan R.K.S., Pike G.B., and Evans A.C., 1997, Brainweb: Online interface to a 3D MRI simulated brain database, NeuroImage 5, 425.
Cohen I., Cohen L., and Ayache N., 1992, Using deformable surfaces to segment 3-d images and infer differential structures, CVGIP 56(2), 242–263.
Cohen L. and Cohen I., 1993, Finite-element methods for active contour models and balloons for 2-d and 3-d images, IEEE Transactions on Pattern Analysis and Machine Intelligence 15(11), 1131–1147.
Cohen L.D., 1991, On active contour models and balloons, Computer Vision Graphics and Image Processing: Image Understanding 53(2), 211–218.
Collette Y. and Siarry P., 2002, Multiobjective Optimization Principles and Case Studies, Decision engineering, Springer, Berlin, Germany.
Cootes T., Taylor C., Cooper D., and Graham J., 1992, Training models of shape from sets of examples, British Machine Vision Conference, pp. 9–18. Springer-Verlag, Leeds, UK.
Cootes T., Taylor C., Hill A., and Halsam J., 1993, The use of active shape models for locating structures in medical images, Proceedings of the 13th International Conference on Information Processing in Medical Imaging, Flagstaff, AZ, pp. 33–47. Springer-Verlag.
Cootes T. and Taylor C., 1997, A mixture model for representing shape variation, British Machine Vision Conference, pp. 110–119. Springer-Verlag, University of Essex, UK.
Cootes T.F., Cooper D., Taylor C.J., and Graham J., 1995, Active shape models—their training and application, Computer Vision and Image Understanding 61, 38–59.
Cootes T.F., Edwards G.J., and Taylor C.J., 2001, Active appearance models, IEEE Transactions on Pattern Analysis and Machine Intelligence 23(6), 681–685.
Cremers D., 2008, Nonlinear dynamical shape priors for level set segmentation, Journal of Scientific Computing 35(2–3), 132–143.
Cremers D., Kohlberger T., and Schnörr C., 2001, Nonlinear shape statistics via kernel spaces, in Pattern Recognition (Proc. DAGM) (eds. Radig B. and Florczyk S.), Vol. 2191 of LNCS, pp. 269–276, Springer, Munich, Germany.
Cremers D., Kohlberger T., and Schnörr C., 2002, Nonlinear shape statistics in Mumford–Shah based segmentation, in Computer Vision—ECCV 2002 (eds. Heyden A. ), Vol. 2351 of LNCS, pp. 93–108, Springer, Copenhagen, Denmark.
Cremers D., Pock T., Kolev K., and Chambolle A., 2011, Convex relaxation techniques for segmentation, stereo and multiview reconstruction, Advances in Markov Random Fields for Vision and Image Processing, MIT Press, Cambridge, MA.
Cremers D., Schmidt F., and Barthel F., 2008, Shape priors in variational image segmentation: Convexity, lipschitz continuity and globally optimal solutions, IEEE Conference on Computer Vision and Pattern Recognition, 2008, CVPR 2008, pp. 1–6, Anchorage, AK.
Dambreville S., Rathi Y., and Tannen A., 2006, Shape-based approach to robust image segmentation using kernel PCA, IEEE Computer Society Conference on Computer Vision and Pattern Recognition, 2006, Vol. 1, pp. 977–984, New York.
Delingette H., 1999, General object reconstruction based on simplex meshes, International Journal of Computer Vision 32, 111–146.
Delingette H., Hebert M., and Ikeuchi K., 1992, Shape representation and image segmentation using deformable surfaces, Image Vision Computing 10(3), 132–144.
Delong A., Osokin A., Isack H., and Boykov Y., 2012, Fast approximate energy minimi-zation with label costs, International Journal of Computer Vision (IJCV) 96(1), 1–27.
Dice L.R., 1945, Measures of the amount of ecologic association between species, Ecology 26(3), 297–302.
Elsgolc L., 1962, Calculus of Variations, Pergamon Press Ltd., London, U.K.
Etyngier P., Segonne F., and Keriven R., 2007, Shape priors using manifold learning techniques, ICCV 2007: IEEE 11th International Conference on Computer Vision, 2007, pp. 1–8, Rio de Janeiro, Brazil.
Falcão A.X., Udupa J.K., Samarasekera S., Sharma S., Hirsch B.E., and Lotufo RdA, 1998, User-steered image segmentation paradigms: Live wire and live lane, Graphical Models and Image Processing 60(4), 233–260.
Falcão A.X. and Udupa J.K., 1997, Segmentation of 3D objects using live wire, in Medical Imaging 1997: Image Processing (ed. Hanson K.M.), Vol. 3034, pp. 228–235. SPIE, Newport Beach, CA.
Fan Y., Jiang T., and Evans D., 2002, Volumetric segmentation of brain images using parallel genetic algorithms, IEEE Transactions on Medical Imaging 21(8), 904–909.
Finley T. and Joachims T., 2008, Training structural SVMs when exact inference is intractable, ICML ‘08: Proceedings of the 25th International Conference on Machine Learning, pp. 304–311, ACM, New York.
Fletcher P., Lu C., Pizer S., and Joshi S., 2004, Principal geodesic analysis for the study of nonlinear statistics of shape, IEEE Transactions on Medical Imaging 23(8), 995–1005.
Frangi A., Niessen W., Hoogeveen R., van Walsum T., and Viergever M., 1999, Quantitation of vessel morphology from 3D MRA, in Medical Image Computing and Computer-Assisted Intervention—MICCAI’99 (eds. Taylor C. and Colchester A.), Vol. 1679 of Lecture notes in Computer Science, pp. 358–367, Springer Berlin/Heidelberg, Germany.
Geoffrion A.M., 1968, Proper efficiency and the theory of vector maximization, Journal of Mathematics, Analysis and Applications 22(3), 618–630.
Ghosh P. and Mitchell M., 2006, Segmentation of medical images using a genetic algorithm, Proceedings of the 8th Annual Conference on Genetic and Evolutionary Computation, GECCO ‘06, pp. 1171–1178, ACM, New York.
Grady L., 2006, Random walks for image segmentation, IEEE Transactions on Pattern Analysis and Machine Intelligence 28(11), 1768–1783.
Grady L. and Funka-Lea G., 2004, Multi-label image segmentation for medical applications based on graph-theoretic electrical potentials, Computer Vision and Mathematical Methods in Medical and Biomedical Image Analysis, pp. 230–245, Prague, Czech Republic.
Grady L. and Polimeni J.R., 2010, Discrete Calculus: Applied Analysis on Graphs for Computational Science, Springer, London, U.K.
Hamarneh G. and Gustavsson T., 2004, Deformable spatio-temporal shape models: extending active shape models to 2D+time, Journal of Image Vision Computing 22(6), 461–470.
Hamarneh G. and Jassi P., 2010, Vascusynth: Simulating vascular trees for generating volumetric image data with ground truth segmentation and tree analysis, Computerized Medical Imaging and Graphics 34(8), 605–616.
Hamarneh G., Jassi P., and Tang L., 2008, Simulation of ground-truth validation data via physically- and statistically-based warps, Lecture Notes in Computer Science, Medical Image Computing and Computer-Assisted Intervention (MICCAI), pp. 459–467.
Hamarneh G., McInerney T., and Terzopoulos D., 2001, Deformable organisms for automatic medical image analysis, in Medical Image Computing and Computer-Assisted Intervention—MICCAI 2001 (eds. Niessen W. and Viergever M.), Vol. 2208 of Lecture notes in Computer Science, pp. 66–76, Springer Berlin/Heidelberg, Germany.
Hamarneh G., McInerney T., and Terzopoulos D., 2001, Deformable organisms for automatic medical image analysis, Lecture Notes in Computer Science, Medical Image Computing and Computer-Assisted Intervention (MICCAI), Vol. 2208, pp. 66–75.
Hamarneh G., McIntosh C., and Drew M., 2011, Perception-based visualization of manifold-valued medical images using distance-preserving dimensionality reduction, IEEE Transactions on Medical Imaging, 30(7), 1314–1327.
Hamarneh G., McIntosh C., McInerney T., and Terzopoulos D., 2009, Deformable organisms: An artificial life framework for automated medical image analysis, in Computational Intelligence in Medical Imaging: Techniques and Applications, CRC Press, Boca Raton, FL, pp. 433–474.
Hamarneh G. and McIntosh C., 2007, Physically and statistically based deformable models for medical image analysis (chapter 11), Deformable Models: Biomedical and Clinical Applications, pp. 335–386, Springer, New York, http://rd.springer.com/chapter/10.1007%2F978-0-387-68413-0_11.
Hanning T., Schöne R., and Pisinger G., 2006, Vector image segmentation by piecewise continuous approximation, Journal of Mathematical Imaging and Vision 25, 5–23, 10.1007/s10851-005-4385-5.
Heimann T. and Meinzer H.P., 2009, Statistical shape models for 3D medical image segmentation: A review, Medical Image Analysis 13(4), 543–563.
Hill A. and Taylor C., 1992, Model-based image interpretation using genetic algorithms, Image and Vision Computing 10(5), 295–300.
Ibáñez O., Barreira N., Santos J., and Penedo M., 2009, Genetic approaches for topological active nets optimization, Pattern Recognition 42(5), 907–917.
Jaccard P., 1901, Étude comparative de la distribution florale dans une portion des alpes et des jura, Bulletin del la Société Vaudoise des Sciences Naturelles 37, 547–579.
Kass M., Witkin A., and Terzopoulos D., 1987, Snakes: Active contour models, International Journal of Computer Vision 1(4), 321–331.
Kawahara J., McIntosh C., Tam R., and Hamarneh G., 2013, Globally optimal spinal cord segmentation using a minimal path in high dimensions, in IEEE International Symposium on Biomedical Imaging, pp. 836–839.
Kolmogorov V., Boykov Y., and Rother C., 2007, Applications of parametric maxflow in computer vision, ICCV 2007: IEEE 11th International Conference on Computer Vision, 2007, pp. 1–8, Rio de Janeiro, Brazil.
Kolmogorov V. and Zabin R., 2004, What energy functions can be minimized via graph cuts? IEEE Transactions on Pattern Analysis and Machine Intelligence 26(2), 147–159.
Lesage D., Angelini E.D., Bloch I., and Funka-Lea G., 2009, A review of 3D vessel lumen segmentation techniques: Models, features and extraction schemes, Medical Image Analysis 13(6), 819–845. Includes Special Section on Computational Biomechanics for Medicine.
Leventon M., Grimson W., and Faugeras O., 2000, Statistical shape influence in geodesic active contours, Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2000, Vol. 1, pp. 316–323, San Diego, CA.
Li H. and Yezzi A., 2006, Vessels as 4d curves: Global minimal 4d paths to extract 3d tubular surfaces, CVPRW ‘06: Conference on Computer Vision and Pattern Recognition Workshop, 2006, pp. 82–82, NY, USA.
Lorigo L.M., Faugeras O.D., Grimson W.E.L., Keriven R., Kikinis R., Nabavi A., and Westin C.F., 2001, Curves: Curve evolution for vessel segmentation, Medical Image Analysis 5(3), 195–206.
MacEachern L. and Manku T., 1998, Genetic algorithms for active contour optimization, ISCAS ‘98: Proceedings of the 1998 IEEE International Symposium on Circuits and Systems, 1998, Vol. 4, pp. 229–232, Monterey, CA.
Maintz J. and Viergever M.A., 1998, A survey of medical image registration, Medical Image Analysis 2(1), 1–36.
McInerney T. and Terzopoulos D., 1995a, A dynamic finite element surface model for segmentation and tracking in multidimensional medical images with application to cardiac 4d image analysis, Computerized Medical Imaging and Graphics 19(1), 69–83.
McInerney T. and Terzopoulos D., 1995b, Medical image segmentation using topologically adaptable snakes, Proceedings of the First International Conference on Computer Vision, Virtual Reality and Robotics in Medicine, pp. 92–101, Springer-Verlag, London, U.K.
McInerney T. and Terzopoulos D., 1995c, Topologically adaptable snakes, Proceedings of the Fifth International Conference on Computer Vision, 1995, pp. 840–845, Cambridge, MA.
McInerney T. and Terzopoulos D., 1996, Deformable models in medical image analysis: A survey, Medical Image Analysis 1(2), 91–108.
McInerney T. and Terzopoulos D., 1999, Topology adaptive deformable surfaces for medical image volume segmentation, IEEE Transactions on Medical Imaging 18(10), 840–850.
McInerney T. and Terzopoulos D., 2000, T-snakes: Topology adaptive snakes, Medical Image Analysis 4(2), 73–91.
McIntosh C., 2011, Energy functionals for medical image segmentation: Choices and consequences, PhD dissertation, Simon Fraser University, Burnaby, BC, Canada.
McIntosh C. and Hamarneh G., 2006a, Vessel crawlers: 3D physically-based deformable organisms for vasculature segmentation and analysis, IEEE Computer Society Conference on Computer Vision and Pattern Recognition, 2006, Vol. 1, pp. 1084–1091, New York.
McIntosh C. and Hamarneh G., 2006b, Genetic algorithm driven statistically deformed models for medical image segmentation, ACM Workshop on Medical Applications of Genetic and Evolutionary Computation Workshop (MedGEC), in conjunction with the Genetic and Evolutionary Computation Conference (GECCO), Seattle, WA.
McIntosh C. and Hamarneh G., 2007, Is a single energy functional sufficient? Adaptive energy functionals and automatic initialization, in Medical Image Computing and Computer-Assisted Intervention—MICCAI 2007 (eds. Ayache N., Ourselin S., and Maeder A.), Vol. 4792 of Lecture notes in Computer Science, pp. 503–510, Springer Berlin/Heidelberg, Germany, 10.1007/978-3-540-75759-7_61.
McIntosh C. and Hamarneh G., 2009, Optimal weights for convex functionals in medical image segmentation, International Symposium on Visual Computing: Special Track on Optimization for Vision, Graphics and Medical Imaging: Theory and Applications (ISVC OVGMI), Vol. 5875-I, pp. 1079–1088, Las Vegas, Nevada, USA.
McIntosh C. and Hamarneh G., 2012, Medial-based deformable models in non-convex shape-spaces for medical image segmentation using genetic algorithms, IEEE Transactions on Medical Imaging, on 31.1, 33–50.
Mignotte M. and Meunier J., 1999, Deformable template and distribution mixture-based data modeling for the endocardial contour tracking in an echographic sequence, IEEE Computer Society Conference on Computer Vision and Pattern Recognition, 1999, Vol. 1, p. 230, Fort Collins, CO.
Miller J.V., Breen D.E., Lorensen W.E., O'Bara R.M., and Wozny M.J., 1991, Geometrically deformed models: A method for extracting closed geometric models form volume data, SIGGRAPH Computer Graphics 25(4), 217–226.
Montagnat J., Delingette H., and Ayache N., 2001, A review of deformable surfaces: Topology, geometry and deformation, Image and Vision Computing 19(14), 1023–1040.
Mortensen E.N. and Barrett W.A., 1998, Interactive segmentation with intelligent scissors, Graphical Models and Image Processing 60(5), 349–384.
Mumford D. and Shah J., 1989, Optimal approximation by piecewise smooth functions and associated variational problems, Communications on Pure Applied Mathematics 42, 577–685.
Nakib A., Oulhadj H., and Siarry P., 2010, Image thresholding based on pareto multiob-jective optimization, Engineering Applications of Artificial Intelligence 23.3: 313–320.
Nand K., Abugharbieh R., Booth B., and Hamarneh G., 2011, Detecting structure in diffusion tensor MR images, Lecture Notes in Computer Science, Medical Image Computing and Computer-Assisted Intervention (MICCAI), pp. 90–97.
Ng B., Hamarneh G., and Abugharbieh R., 2012, Modeling brain activation in fMRI using group MRF, IEEE Transactions on Medical Imaging 31(5), 1113–1123.
Nikolova M., Esedoglu S., and Chan T.F., 2006, Algorithms for finding global mini-mizers of image segmentation and denoising models, SIAM Journal on Applied Mathematics 66(5), 1632–1648.
Nosrati M. and Hamarneh G., 2013, Segmentation of cells with partial occlusion and part configuration constraint using evolutionary computation, Lecture Notes in Computer Science, Medical Image Computing and Computer-Assisted Intervention (MICCAI).
O'Donnell T., Boult T., Fang X.S., and Gupta A., 1994, The extruded generalized cylinder: A deformable model for object recovery, Proceedings CVPR ‘94: IEEE Computer Society Conference on Computer Vision and Pattern Recognition, 1994, pp. 174–181, Seattle, WA.
Osher S. and Paragios N., 2003, Geometric Level Set Methods in Imaging Vision and Graphics, Springer Verlag, New York.
Osher S. and Sethian J.A., 1988, Fronts propagating with curvature dependent speed: Algorithms based on Hamilton-Jacobi formulations, Journal of Computational Physics 79(1), 12–49.
Paragios N. and Deriche R., 2000, Coupled geodesic active regions for image segmentation: A level set approach, in Computer Vision—ECCV 2000 (eds. Vernon D.), Vol. 1843 of Lecture notes in Computer Science, pp. 224–240, Springer Berlin/Heidelberg, Germany.
Paragios N. and Deriche R., 2002, Geodesic active regions and level set methods for supervised texture segmentation, International Journal of Computer Vision 46, 223–247.
Paragios N., Taron M., Huang X., Rousson M., and Metaxas D., 2006, On the representation of shapes using implicit functions, in Statistics and Analysis of Shapes (eds. Krim H. and Yezzi A.), Modeling and simulation in science, engineering and technology, Birkhäuser, Boston, MA, pp. 167–199.
Pham D.L., Xu C., and Prince J.L., 2000, A survey of current methods in medical image segmentation, In Annual Review of Biomedical Engineering 2, 315–338.
Pizer S.M., 2003, Guest editorial—Medial and medical: A good match for image analysis, International Journal of Computer Vision 55(2–3), 79–84.
Pizer S.M., Fletcher P.T., Joshi S.C., Thall A., Chen J.Z., Fridman Y., Fritsch D.S. , 2003, Deformable M-Reps for 3d medical image segmentation, International Journal of Computer Vision 55(2–3), 85–106.
Pohl K.M., Fisher J., Bouix S., Shenton M., McCarley R.W., Grimson W.E.L., Kikinis R., and Wells W.M., 2007, Using the logarithm of odds to define a vector space on probabilistic atlases, Medical Image Analysis 11(5), 465–477, Special issue on the Ninth International Conference on Medical Image Computing and Computer-Assisted Interventions—MICCAI 2006.
Poon M., Hamarneh G., and Abugharbieh R., 2008, Efficient interactive 3D livewire segmentation of objects with arbitrarily topologies, Computerized Medical Imaging and Graphics 32(8), 639–650.
Rana M., Hamarneh G., and Wakeling J., 2009, Automated tracking of muscle fascicle orientation in B-mode ultrasound images, Journal of Biomechanics 42(13), 2068–2073.
Rao J., Abugharbieh R., and Hamarneh G., 2010, Adaptive regularization for image segmentation using local image curvature cues, European Conference on Computer Vision (ECCV), pp. 651–665.
Rao J., Hamarneh G., and Abugharbieh R., 2009, Adaptive contextual energy parameterization for automated image segmentation, in Lecture Notes in Computer Science, International Symposium on Visual Computing: Special Track on Optimization for Vision, Graphics and Medical Imaging: Theory and Applications (ISVC OVGMI), Vol. 5875-I, pp. 1089–1100.
Robb R.A., 2000, Biomedical Imaging, Visualization, and Analysis, Wiley-Liss Inc., New York.
Rousson M., Lenglet C., and Deriche R., 2004, Level set and region based surface propagation for diffusion tensor MRI segmentation, in Computer Vision and Mathematical Methods in Medical and Biomedical Image Analysis (eds. Sonka M., Kakadiaris I.A., and Kybic J.), Vol. 3117 of Lecture notes in Computer Science, pp. 123–134, Springer Berlin/Heidelberg, Germany, 10.1007/978-3-540-27816-0_11.
Ruff C.F., Hughes S.W., and Hawkes D.J., 1999, Volume estimation from sparse planar images using deformable models, Image and Vision Computing 17(8), 559–565.
Saad A., Hamarneh G., and Moeller T., 2010a, Exploration and visualization of segmentation uncertainty using shape and appearance prior information, IEEE Transactions on Visualization and Computer Graphics (Special Issue of the IEEE Visualization Conference 2010) 16(6), 1366–1375.
Saad A., Hamarneh G., Moeller T., and Smith B., 2008, Kinetic modeling based probabilistic segmentation for molecular images, Lecture Notes in Computer Science, Medical Image Computing and Computer-Assisted Intervention (MICCAI), pp. 244–252.
Saad A., Moeller T., and Hamarneh G., 2010b, ProbExplorer: An uncertainty-based visual analysis tool for medical imaging, Computer Graphics Forum (CGF) (Special Issue of the proceedings of Eurographics/IEEE-VGTC Symposium on Visualization 2010) 29(3), 1113–1122.
Samson C., Blanc-Féraud L., Aubert G., and Zerubia J., 2000, A level set model for image classification, International Journal of Computer Vision 40, 187–197, 10.1023/A:1008183109594.
Sapiro G., 1996, Vector-valued active contours, Proceedings CVPR ‘96: IEEE Computer Society Conference on Computer Vision and Pattern Recognition, 1996, pp. 680–685, San Francisco, CA, USA.
Sethian J., 1996, Level Set Methods: Evolving Interfaces in Geometry, Fluid Mechanics, Computer Vision and Materials Science, Cambridge University Press, Cambridge, U.K.
Shah J., 1996, Curve evolution and segmentation functionals: Application to color images Proceedings of the International Conference on Image Processing, 1996, Vol. 1, pp. 461–464, Lausanne, Switzerland.
Shi J. and Malik, J., 2000, Normalized cuts and image segmentation, Pattern Analysis and Machine Intelligence, IEEE Transactions on 22.8: 888–905.
Shi L., Funt B., and Hamarneh G., 2008, Quaternion color curvature, Color Imaging (IS&T/SID CI), pp. 338–341, Portland, Oregon.
Sinop A. and Grady L., 2007, A seeded image segmentation framework unifying graph cuts and random walker which yields a new algorithm, ICCV 2007: IEEE 11th International Conference on Computer Vision, 2007, pp. 1–8, Rio de Janeiro, Brazil.
Sonka M. and Fitzpatrick J., 2000, Handbook of Medical Imaging, Volume 2: Medical Image Processing and Analysis, SPIE-International Society for Optical Engine, http://spie.org/X648.html?produce_id_=_831079
Sozou P., Cootes T., Taylor C., and Di Mauro E., 1995, Non-linear point distribution modelling using a multi-layer perceptron, British Machine Vision Conference, pp. 107–116, Birmingham, U.K.
Staib L. and Duncan J., 1992a, Boundary finding with parametrically deformable models, IEEE Transactions on Pattern Analysis and Machine Intelligence 14(11), 1061–1075.
Staib L.H. and Duncan J.S., 1992b, Deformable Fourier models for surface finding in 3D images, Proceedings of the Second Conference on Visualization in Biomedical Computing (VBC-92), Vol. 1808, pp. 90–104, SPIE, Bellingham, WA.
Sundaramoorthi G., Yezzi A., and Mennucci A.C., 2007, Sobolev active contours, International Journal of Computer Vision 73(3), 345–366.
Szummer M., Kohli P., and Hoiem D., 2008, Learning CRFs using graph cuts, in Computer Vision—ECCV 2008 (eds. Forsyth D., Torr P., and Zisserman A.), Vol. 5303 of Lecture notes in Computer Science, pp. 582–595, Springer Berlin/Heidelberg, Germany.
Tang L., Bressmann T., and Hamarneh G., 2012, Tongue contour tracking in dynamic ultrasound via higher-order MRFs and efficient fusion moves, Medical Image Analysis 16(8), 1503–1520.
Tanimoto T., 1957, An elementary mathematical theory of classification and prediction, Technical report, IBM Internal Report.
Taskar B., Chatalbashev V., Koller D., and Guestrin C., 2005, Learning structured prediction models: A large margin approach, ICML ‘05: Proceedings of the 22nd International Conference on Machine Learning, pp. 896–903, ACM, New York.
Terzopoulos D., 1987, On matching deformable models to images, Topical Meeting on Machine Vision, Technical Digest Series 12, 160–167.
Tohka J., 2001, Global optimization of deformable surface meshes based on genetic algorithms, Proceedings of the 11th International Conference on Image Analysis and Processing, 2001, pp. 459–464, Palermo, Italy.
Top A., Hamarneh G., and Abugharbieh R., 2010, Spotlight: Automated confidence-based user guidance for increasing efficiency in interactive 3D image segmentation, in Medical Image Computing and Computer-Assisted Intervention Workshop on Medical Computer Vision (MICCAI MCV), Vol. 6533, pp. 204–213.
Top A., Hamarneh G., and Abugharbieh R., 2011, Active learning for interactive 3D image segmentation, in Lecture Notes in Computer Science, Medical Image Computing and Computer-Assisted Intervention (MICCAI), Vol. 6893, pp. 603–610.
Tsai A., Wells W., Tempany C., Grimson E., and Willsky A., 2004, Mutual information in coupled multi-shape model for medical image segmentation, Medical Image Analysis 8(4), 429–445.
Tsai A., Yezzi A.J., and Willsky A., 2001, Curve evolution implementation of the Mumford-Shah functional for image segmentation, denoising, interpolation, and magnification, IEEE Transactions on Image Processing 10(8), 1169–1186.
Ulen J., Strandmark P., and Kahl F., 2013, An efficient optimization framework for multi-region segmentation based on Lagrangian duality, IEEE Transactions on Medical Imaging 32(2), 178–188.
Vasilevskiy A. and Siddiqi K., 2002, Flux maximizing geometric flows, IEEE Transactions on Pattern Analysis and Machine Intelligence 24(12), 1565–1578.
Vese L.A. and Chan T.F., 2001, A multiphase level set framework for image segmentation using the Mumford and Shah model, International Journal of Computer Vision 50, 271–293.
Vu N. and Manjunath B., 2008, Shape prior segmentation of multiple objects with graph cuts, CVPR 2008: IEEE Conference on Computer Vision and Pattern Recognition, 2008, pp. 1–8, Anchorage, AK.
Wang Z. and Vemuri B.C., 2004, Tensor field segmentation using region based active contour model, Computer Vision—ECCV 2000, Vol. 3024 of Lecture notes in Computer Science, pp. 304–315, Springer Berlin/Heidelberg, Germany.
Warfield S., Kaus M., Jolesz F.A., and Kikinis R., 2000, Adaptive, template moderated, spatially varying statistical classification, Medical Image Analysis 4(1), 43–55.
Warfield S., Zou K., and Wells W., 2004, Simultaneous truth and performance level estimation (staple): An algorithm for the validation of image segmentation, IEEE Transactions on Medical Imaging 23(7), 903–921.
Weldeselassie Y. and Hamarneh G., 2007, DT-MRI segmentation using graph cuts, SPIE Medical Imaging, Vol. 6512-1K, pp. 1–9, San Diego, California, USA.
Whitaker R.T., 1994, Volumetric deformable models: Active blobs, Visualization in Biomedical Computing 2359(1), 122–134.
Wink O., Niessen W., Verdonck B., and Viergever M., 2001, Vessel axis determination using wave front propagation analysis, in Medical Image Computing and Computer-Assisted Intervention—MICCAI 2001 (eds. Niessen W. and Viergever M.), Vol. 2208 of Lecture notes in Computer Science, pp. 845–853, Springer, Berlin/Heidelberg, Germany, 10.1007/3-540-45468-3_101.
Wink O., Niessen W.J., and Viergever M., 2004, Multiscale vessel tracking, IEEE Transactions on Medical Imaging 23(1), 130–133.
Xu C. and Prince J., 1998, Snakes, shapes, and gradient vector flow, IEEE Transactions on Image Processing 7(3), 359–369.
Yazdanpanah A., Hamarneh G., Smith B., and Sarunic M., 2011, Automated segmentation of intra-retinal layers from optical coherence tomography images using an active contour approach, IEEE Transactions on Medical Imaging 2(30), 484–496.
Yezzi A.J., Kichenassamy S., Kumar A., Olver P., and Tannenbaum A., 1997, A geometric snake model for segmentation of medical imagery, IEEE Transactions on Medical Imaging 16(2), 199–209.
Yoo T.S., 2004, Insight into Images: Principles and Practice for Segmentation, Registration, and Image Analysis, A K Peters Ltd., Wellesey, MA.
Zhao H.K., Chan T., Merriman B., and Osher S., 1996, A variational level set approach to multiphase motion, Journal of Computational Physics 127(1), 179–195.
Zitová B. and Flusser J., 2003, Image registration methods: A survey, Image and Vision Computing 21(11), 977–1000.
* This chapter summarizes and builds upon McIntosh (2012).
Index
A
Aberrant crypt foci (ACF), 62, 64–66
ABF, see Analog beamforming (ABF)
ACF, see Aberrant crypt foci (ACF)
Acquisition electronics, 372–376
ACS, see Autocalibration signal (ACS)
ACT, see Average CT (ACT)
Active shape models (ASM), 671, 675
AD, see Alzheimer's disease (AD)
ADC, see Analog-to-digital converters (ADCs)
Algebraic reconstruction technique (ART), 132
Alzheimer's disease (AD), 583–584
Analog beamforming (ABF), 223–224
Analog-to-digital converters (ADCs)
CAD tools, 237
output signals, 278
PET and SPECT designs, 375
power efficiency vs. dynamic range, 227
range and bandwidth, 226
SSV technique, 237
TIQ comparator models, accuracy comparison, 239
transistor yields, 241
Application-specific integrated circuits (ASICs), 122, 263, 313
ART, see Algebraic reconstruction technique (ART)
ASICs, see Application-specific integrated circuits (ASICs)
ASM, see Active shape models (ASM)
Attenuation map, creation, 343–348
Autocalibration signal (ACS), 499, 506
Avalanche photodiode (APD)
breakdown voltage, 383
representation, 382
electrical signals, 386
Geiger mode, 386
quenching, 386
radiation detectors, 383
spatial resolution, 384
system gain, 383
Average CT (ACT), 174, 185, 186
B
Bayesian network (BN) modeling
directed Markov property and faithfulness assumption, 604
joint probability distribution, 603, 604
PCfdr algorithm, see PCfdr algorithm
BDL, see Biliary duct ligation (BDL)
Beamforming
post processor adds, 223
PPL, 224
receive beamformer, 223
transmit beamformer, 223
ultrasound images, finer resolution, 224
Beam-hardening, 344
Biliary duct ligation (BDL), 584–585
Block-matching (BM) algorithm, 646
Blood oxygen level dependent (BOLD)
description, 594
DTI, 595
functional connectivity, 596
GLM, 596
ICA, 599
seed-based ROI method, 597
T2-weighted contrasts, 570
BM algorithm, see Block-matching (BM) algorithm
B-mode imaging system hardware
A/D converter and memory (FIFO), 222–223
amplifier, 222
block diagram, 222
control host, 221
TGC, 222
T/R switch, 222
BN modeling, see Bayesian network (BN) modeling
BOLD, see Blood oxygen level dependent (BOLD)
Bone and soft-tissue maps, 344
Brain connectivity
diffusion MRI, see Diffusion magnetic resonance imaging (dMRI)
functional MRI
assessment, network modeling methods, 601
behavioral paradigms, 595
BN modeling, see Bayesian network (BN) modeling
BOLD, see Blood oxygen level dependent (BOLD)
clustering methods, 599
conditional-dependence measures, 598–599
connectivity network, ROIs, 612–613
graphical models, 601
incrementally updatable and hierarchical group analysis, 613
linear decomposition methods, 599
multivariate statistical methods, 600
perturbation analysis, 614
seed-based ROI method, 597
temporal and spatial
resolution, 594
Breathing instruction-based methods, PET/CT
normal end-expiration breath-hold, 407–408
shallow breathing, 407
C
CA algorithms, see Coordinate accent (CA) algorithms
CACT, see Cine average CT (CACT)
CAD, see Computer aided design (CAD)
Cadmium telluride (CdTe), detectors, 388–390
Cadmium zinc telluride (CdZnTe), detectors, 388–390
Calibration-Less Multicoil (CaLM) MRI
conditions, CS recovery, 514
data acquisition model, 513–514
frequency domain methods, 513
Gaussian sampling, 517
group-sparse reconstruction, 514–516
reconstruction accuracy
brain data and Shepp–Logan phantom, 517–518
GRAPPA, l1SPIRiT and CS SENSE, 519, 521
NMSE, see Normalized mean squared error (NMSE)
objectives, 518
CaLM MRI, see Calibration-Less Multicoil (CaLM) MRI
Capacitive micromachined ultrasonic transducers (CMUTs) imaging systems
applications
large/high-density 2D array, 266–268
in vivo, 263
benefits
grating lobes concept, 259–260
IC integration, 260
cell, cross-sectional schematic, 255
challenges
efficiency, 269
reliability, 269
transducer elements, cross talk, 269
CMUT–CMOS integration techniques
ASICs, 263
CMUT-in-CMOS, 261
flip-chip integration, 261–262
design and fabrication
and application, 254
cavities formation, 257
horizontal dimension, 256
integrated circuit (IC) production, 255
silicon nitride and silicon oxide, 257
square-and hexagon-shaped membranes, 256
MEMS-based transducer, 254
scanning electron microscope image, 254
ultrasonic transducers composition, 254
Cascaded system theory (CST), 94
CCD, see Charge-coupled devices (CCD)
CEST imaging, see Chemical exchange saturation transfer (CEST) imaging
CFD, see Constant fraction discriminator (CFD)
Charge-coupled devices (CCD), 110, 113, 140
Charge-sensitive preamplifier (CSA), 372–374
CHARMED, see Composite hindered and restricted model of diffusion (CHARMED)
Chemical exchange saturation transfer (CEST) imaging, 571–572
CIE color functions, see Commission Internationale d'Eclairage (CIE) color functions
Cine average CT (CACT), 411–413
Closed-form solutions, MIR
affine transformations, 636
least squares (LS) error, 634–635
nonlinear transform, 634
spectral algorithms, 636
Clustering methods, brain connectivity, 599
CMOS, see Complementary metal oxide semiconductor (CMOS)
CMUTs imaging systems, see Capacitive micromachined ultrasonic transducers (CMUTs) imaging systems
CNR, see Contrast-to-noise ratio (CNR)
Commercial helical 4DCT systems, 182–183
Commission Internationale d'Eclairage (CIE) color functions, 73, 74
Compartmental model
receptor-ligand binding
DVR, 470
equilibrium dissociation constant, 468
radioligand concentration, plasma, 469
single-tissue compartment, 470
Complementary metal oxide semiconductor (CMOS), 118, 119, 125, 130
Composite hindered and restricted model of diffusion (CHARMED), 542–543
Compressed sensing (CS), MRI acceleration
description, 485
estimation, sparse vector, 486
hardware and software based approaches, 485–486
multichannel scans
calibration-free parallel MRI reconstruction, see Calibration-Less Multicoil (CaLM) MRI
data acquisition process, 499
k-space samples, 499
SENSE-based reconstruction, 500–513
SMASH, GRAPPA and ACS, 499
single-channel MRI scans
data acquisition model, 488
k-space, 488
low-rank matrix, see Low-rank matrix, MRI reconstruction
Computed tomography (CT)
iterative methods, 210
protocols
low-pitch CT, 411
scan modes, 410
Computer aided design (CAD), 237
Cone-beam
artifacts, 212
reconstruction
description, 204
fan-to-parallel-beam rebinning method, 206
FDK algorithm, 205
hybrid filtering approach, 205–206
image artifacts, 207
phantom, 207
Wedge algorithm, 206
Constant fraction discriminator (CFD), 279
Contrast-to-noise ratio (CNR)
dual-energy imaging, 153
lesion detection, 155
performance, 303
Coordinate accent (CA) algorithms, 441–442
Corticospinal fluid (CSF), 531
Crossed electrodes, see Row-column (RC)
CSA, see Charge-sensitive preamplifier (CSA)
CSF, see Corticospinal fluid (CSF)
CSI, see Cubic spline interpolation (CSI)
CST, see Cascaded system theory (CST)
CT, see Computed tomography (CT)
CT-based co-registration, 361–362
CT protocols, see Computed tomography (CT)
CT-SPECT/CT-PET
attenuation
beam-hardening, 344
blurred image, 344
bone and soft-tissue maps, 344
CT vs. transmission scans, 338–339
Hounsefield units, 343
max-intensity CT, 348
metal objects, 345
patient movement, 345
CTAC
FDG PET/CT whole-body study, 340
field-of-view (FOV), 339
photon flux, 339
radioactive sources, 339
speed, 341
diagnostic information
calcium scoring, 348
high-resolution anatomical map, 348–349
non-oncologic studies, 348
radiotracers, 349
image registration
motion compensation, areas, 351
nonlinear registration algorithms, 351
rigid-body registration, 349
semi-automated methods, 349
software registration, 349–351
warping algorithms, 350
localization advantages
bone scan, 338
cancer, focal sites, 337
cardiac imaging, 338
DEFER trial, 338
nuclear medicine, 337
MRI
absolute quantification, 352
cost-effectiveness, 353
metal implants, 353
patient-specific attenuation, 353
photomultiplier tubes, 352
radiofrequency radiation, 351–352
solid-state amplification, 352
multislice scanners, 336
“unclear medicine”, 335
Cubic spline interpolation (CSI), 40–41
CZT, see Cadmium zinc telluride (CdZnTe), detectors
D
DAG, see Directed acyclic graph (DAG)
Data acquisition, micro-CT systems
calculation, 130
continuous rotation, 128
detector integration period, 129
online correction, raw data, 130–131
projection images, 129
step-and-shoot approach, 128
Data interpolation, MIR, 621–622
Data sufficiency condition (DSC), 176–178
DBF, see Digital beamforming (DBF)
DCT, see Discrete cosine transformation (DCT)
4DCT, see Four-dimensional computed tomography (4DCT)
Deep-inspiration breath-hold (DIBH) PET/CT, 409–410
Demons algorithm, MIR
Gaussian smoothing, 643
modification, 645
orientation and magnitude, forces, 643
PASHA algorithm, 644
Depth-of-interaction (DoI)
detector design concepts, 315
dual-layer crystals, offset positions, 316–317
monolithic crystals, statistical positioning, 316
multiple crystal-photodetector layers, 314
Phoswich design, 315
single-crystal layer/dual-ended photodetectors, 314–315
TRI and RECT crystal, 317
Detector response function (DRF)
counting direct conversion detectors, 103–105
indirect and direct conversion detector, 101–102
integrating indirect conversion detectors, 102–103
spectral behavior, 102
DFT, see Discrete Fourier transformation (DFT)
DIBH PET/CT, see Deep-inspiration breath-hold (DIBH) PET/CT
Diffusion magnetic resonance imaging (dMRI)
anisotropy, tissue, 531
brain connectivity mapping
front propagation tractography, 553–556
probabilistic tractography, 550–553
streamline tractography, 548–550
Brownian motion, 530
CSF, gray matter and white matter, 531
diffusion tensor, see Diffusion tensor
HARDI, see Higher angular resolution diffusion imaging (HARDI)
molecular displacement, 530
noise reduction, 547
spin property, magnetic moment, 531–532
subject motion, 536
Diffusion ODF, see Diffusion orientation distribution function (ODF)
Diffusion orientation distribution function (ODF)
DOT, 544
q-ball imaging, 543
tractosema, 548
Diffusion orientation transform (DOT), 544
Diffusion tensor
Brownian motion, PDF, 537
ellipsoid, 539
fitting procedure, 538
mean diffusivity (MD) and fractional anisotropy (FA), 539–541
q-space imaging, 537
quality, 538
visualization strategy, 540, 541
zero-mean Gaussian, 538
Diffusion tensor imaging (DTI), 595, 613
Diffusion-weighted imaging (DWI)
brain's white matter, 534, 535
gradient direction, 535
HARDI, see Higher angular resolution diffusion imaging (HARDI)
magnetic pulse, spin direction, 533
Stejskal–Tanner, 533
subject motion, 536
T2 relaxation times, 534
Digital beamforming (DBF), 223–224
Digital signal interfacing, 225
Digital SiPM
ADC and TDC functions, 324
CMOS
process, 323
dark count rate (DCR), 323
quenching, 323
Direct conversion detector
cathode CZT, 92
flat-panel X-ray
ASIC, 122
cadmium telluride (CdTe), 121
crystallization and stabilize material, 121
design, contact plates, 121
photoconductor materials, 119–120
TFTs, 122
parameters, 94
primary energy deposition, 96
signal conversion steps, 92, 93
Direct conversion detectors, 297–298
Directed acyclic graph (DAG), 603–604, 606, 607
Discrete cosine transformation (DCT), 625
Discrete Fourier transformation (DFT), 625
Discrete optimization, see Markov Random Field (MRF) energy minimization
Distribution volume ratio (DVR), 470
Dixon's composite spin-lock pulse and phase cycling, 577–578
dMRI, see Diffusion magnetic resonance imaging (dMRI)
DoI, see Depth-of-interaction (DoI)
DOT, see Diffusion orientation transform (DOT)
DRF, see Detector response function (DRF)
DSC, see Data sufficiency condition (DSC)
DTI, see Diffusion tensor imaging (DTI)
Dual-energy CT imaging
acquisition, see Image acquisition
description, 147
patient motion, 149
post-contrast single-energy abdomen CT exam, 148
reconstruction, see Image reconstruction, dual-energy x-ray CT imaging
“rotate–rotate” technique, 149
visualization and clinical applications
beam-hardening reduction, 168–169
hepatic cyst, 166
histograms/scatterplots, 167
metal artifact reduction, 169
Dual-layer crystals, offset positions, 316–317
Dual window (DW) processing method
depth resolved spectra, 57, 58
Gaussian distribution, 57
local oscillations, 58
time-frequency distribution (TFD), 56–57
windowed data, 56
DVR, see Distribution volume ratio (DVR)
DWI, see Diffusion-weighted imaging (DWI)
Dynamic PET data model, 452–453
E
EAs, see Evolutionary algorithms (EAs)
Electronics, PET and SPECT imaging
ADC, 375
bias capacitor, voltage, 373
event location, 375
gamma ray deposition, 376
high count rates, measurement, 375
multiple modular detector blocks, 376
output voltage, 374
photocurrent, 372
preamplifier (pre-amp), 372
EM algorithm, see Expectation maximization (EM) algorithm
EMPIRE, see Evaluation of Methods for Pulmonary Image Registration (EMPIRE)
Energy functions, MIS
challenges, 679
description, 664
GACs, 666
graph cuts, 674
noise, 666
parameters, 668
piecewise-constant functions, 667
shape-based terms, 667
ENF, see Excess noise factor (ENF)
Euler–Lagrange equation, 673, 680–681
Evaluation of Methods for Pulmonary Image Registration (EMPIRE), 649–650
Evolutionary algorithms (EAs), 639–640
Excess noise factor (ENF), 289
Expectation maximization (EM) algorithm
closed-form solution, 446
E-step, 443
generalized EM (GEM), 445
pair-wise penalty, 445
Poisson log-likelihood function, 444
EyeCam
camera, 4
durability, 3
person's eyesight, 2
size, power and heat, 3
F
FACT approach, see Fiber assignment by continuous tracking (FACT) approach
Fan-beam projections
analytical methods, 197
backprojection process, 200
divergent rays, 200
kernel hR shift-invariant, 198
and parallel-beam projections, 197, 198
pin-hole and divergent collimators, 197
ramp filter kernel, 199
Fan-to-parallel-beam rebinning method, 206
Fast-kVp switching (FKS), see Dual-energy CT imaging
FDA, see Food and Drug Administration (FDA)
FDK algorithm, see Feldkamp algorithm (FDK) algorithm
FD-OCT, see Fourier domain OCT (FD-OCT)
Feldkamp algorithm (FDK) algorithm, 205
FFD, see Free-form deformation (FFD)
18F–fluorodeoxyglucose (FDG) imaging, 364
Fiber assignment by continuous tracking (FACT) approach, 549, 550
Fiber-optic coupling
limitations, 380
McPET II, 380
timing resolution, 380
Field-programmable gate array (FPGA)
chip, 229
FD-OCT image reconstruction, 25
Flat-panel X-ray detectors
Argus PET/CT, 124
classification, 114
CMOS, 125
edge response function (ERF), 134, 135
features, 124
Hamamatsu C7940DK-02, 124
MTF and, 135
protocol, measurement, 133–134
scintillator layer, 125
slanted-edge method, 134
small-animal imaging, 114
temporal instability, 133
fLCI, see Fourier domain low coherence interferometry (fLCI)
Food and Drug Administration (FDA), 578
Four-dimensional computed tomography (4DCT)
commercial helical 4DCT systems, 182–183
data acquisition modes, 178, 179
image location, slice thickness and scan time, 178, 180
LightSpeed MSCT scanner, 174, 175
respiratory monitoring devices, 183–184
scan modes, 176
Siemens Somatom 4-slice CT, 174–175
work flow and phase selection accuracy, 180–182
Fourier domain low coherence interferometry (fLCI)
animal experimental protocols, 63
depth resolved DW spectra, 60–62
ex vivo hamster cheek pouch carcinogenesis model., 62
measurement, 58
neoplastic development, 66
nuclear morphology measurements, 64, 66
phantom, 60
polystyrene microsphere, 60
scattering properties, 59
statistical analysis, 64
Fourier domain OCT (FD-OCT)
A-scan depth-resolved imaging, 30–31
data processing
numerical dispersion compensation, 41–42
numerical interpolation method, see Numerical interpolation method
imaging resolution and depth, 31, 32
spectrometer-based spectral domain, 29
swept-laser-based swept-source, 29
system design
data processing flow, 38
Fourier rebinning
FOREX algorithm, 434
Fourier slice theorem, 191–193
FPGA, see Field-programmable gate array (FPGA)
FPGA chip, see Field-programmable gate array (FPGA)
Free-form deformation (FFD), 624–625, 631, 642
Front propagation tractography
description, 553
fast marching tractography, 553–554
minimal path algorithms, 555–556
Full width half maximum (FWHM), 302
Functional imaging technique, 360
FWHM, see Full width half maximum (FWHM)
G
GACs, see Geodesic active contours (GACs)
GAPD cells, see Geiger-mode avalanche photodiode (GAPD) cells
GAs, see Genetic algorithms (GAs)
Gated four-dimensional (4D) PET/CT
breathing motions and 18F-FDG uptake, 414
description, 423
distances, lesion centroids, 415, 416
reconstruction-based methods
advantage, 420
deformation matrices, 419
motion compensation, 418
OPL-EM algorithm, 419
registration-based methods
frames, 416
RPM marker location and electrical triggers, 414, 415
Geiger-mode avalanche photodiode (GAPD) cells, 292–293
Generalized autocalibrating partially parallel acquisitions (GRAPPA)
description, 507
image reconstruction
brain, 510
iSENSE CS vs. iSENSE NN, 508–509
Generalized least squares (GLS), 474, 477
Generalized weighted least squares (GWLS), 474, 477
General linear model (GLM), 596
General-purpose computing on graphics processing units (GPGPU), 25, 26
Genetic algorithms (GAs), 674–675
Geodesic active contours (GACs), 666, 678
GLM, see General linear model (GLM)
GLS, see Generalized least squares (GLS)
GPGPU, see General-purpose computing on graphics processing units (GPGPU)
Gradient-based algorithms
Hessian matrix, 440
Newton–Raphson method, 439
PCG algorithm, 440
Gradient-based optimization
gradient descent method, 637
Kiefer–Wolfowitz method, 638
QN and NCG methods, 638
GRAPPA, see Generalized autocalibrating partially parallel acquisitions (GRAPPA)
Group modeling, brain connectivity, 601–602
GWLS, see Generalized weighted least squares (GWLS)
H
Halfscan artifacts, 213
Half-value thickness (HVT), 338
HAMMER, see Hierarchical Attribute Matching Mechanism for Elastic Registration (HAMMER)
Health care
digital and medical imaging
companies and industry, 7
evolution, 5
fusion, multiple imaging modalities, 6
migration to digital files, 5
moving, diagnostic to therapeutic, 5–6
real-time processing, 5
ultrasound, 6
wireless connectivity, 6
life-changing application, medical technology, 4
medical devices, 4
Hierarchical Attribute Matching Mechanism for Elastic Registration (HAMMER), 634, 647
Higher angular resolution diffusion imaging (HARDI)
CHARMED and FORECAST methods, 542–543
DOT, 544
limitations, 543
neural pathways, 542
PAS approach, 544
Stejskal–Tanner equation, 542
High photon detection sensitivity
dual-panel CZT-based PET system, 305–306
geometric efficiency, 304
intrinsic coincidence detection efficiency, 304
rectangular PET detectors and, 306
High-voltage excitation pulses, 225
Hounsfield units (HU), 343, 364
HU, see Hounsfield units (HU)
HVT, see Half-value thickness (HVT)
I
IACT, see Interpolated average CT (IACT)
ICA, see Independent component analysis (ICA)
ICE, see Intracardiac echocardiography (ICE)
ICS, see Intercrystal scatter (ICS)
ICs, see Integrated circuits (ICs)
Image acquisition
dose
region of interest (ROI), 156–157
flux
CNR, 153
modulating, tube current, 154
noise reduction processing, 152
simulated low-contrast lesion, 155
x-ray exposure, 153
Image co-registration, 362
Image reconstruction (IR)
central slice theorem, 191–193
cone-beam reconstruction, see Cone-beam
3D PET
dynamic PET data model, 452–453
Fourier transform, parallel projections, 430–432
linear kinetic models, 454–455
nonlinear kinetic models, 456
optimization algorithms, see Optimization algorithms
2-D Radon transform, 190
dual-energy x-ray CT imaging
effective atomic number, 162–164
energy-dependent attenuation measurements, 159
projection-space material decomposition, 158–159
fan-beam projections, see Fan-beam projections
Hilbert transform, 211
iterative reconstruction methods, see Iterative image reconstruction
parameters, 190
redundancy
conjugate ray, 201
electrocardiogram-gated cardiac image, 203
fan-beam projections, 203
halfscan algorithm, 202
primary ray, 201
scan / partial scan, 203
spatial distribution, 189
trans-axial truncation, 211
Improved spatial resolution
anger-logic PET block detector, 311
complex and expensive assembly, 308–309
compton scatter and multiple interactions, 310
high-density data acquisition systems, 312–313
high-resolution PET detector developments, 312
MTF, 307
PET image quality, 307
PSF, 307
reduced scintillation light output, 309–310
Rose criterion, 308
spatial decoding, block detectors, 310–312
Independent component analysis (ICA), 597, 599
Indirect conversion detectors
CT scintillation detector, 91
electronic read-out, 96
flat-panel X-ray
light transport, nonstructured scintillator screen, 117
optical photons, 115
scintillation materials, 115, 116
secondary quanta detection, 118–119
semiconductor detector, 117
light transport and detection, 96
parameters, 94
primary energy deposition, 94, 96
signal conversion steps, 92, 93
111In-labeled imaging, 365
Integrated circuits (ICs), 1, 11, 13, 21
Intercrystal scatter (ICS), 282–283
Interpolated average CT (IACT), 413–414
Intracardiac echocardiography (ICE), 263–264
Intravascular ultrasound (IVUS), 263–264
IR, see Image reconstruction (IR)
iSENSE method
iSENSE CS vs. iSENSE NN, 508–509
reconstruction
SparSENSE and NNSENSE, 507
Iterative image reconstruction
CT, 210
IVUS, see Intravascular ultrasound (IVUS)
J
Joint image reconstruction and sensitivity estimation in SENSE (JSENSE)
Chebyshev polynomials, 502
CS and nuclear norm minimization, 504–506
iSENSE CS vs. iSENSE NN, 508–509
reconstructed and difference images
brain, 510
JSENSE, see Joint image reconstruction and sensitivity estimation in SENSE (JSENSE)
L
Large/high-density 2D array
LCD, see Low-contrast detectability (LCD)
LDDD, see Log Domain Diffeomorphic Demons (LDDD)
Light scattering spectroscopy (LSS), 59, 60, 62
Linear kinetic models, 454–455
Linear least squares (LLS)
description, 474
single-tissue compartment models
dynamic H215O PET, 478
LWLS and GWLS, 477
measurements, 476
time–activity curves, 476, 478
two-tissue compartment models
Linear spline interpolation (LSI), 39–40
LLS, see Linear least squares (LLS)
LNA, see Low-noise amplifier (LNA)
Log Domain Diffeomorphic Demons (LDDD), 645–646
Low-contrast detectability (LCD), 155–157
Low-noise amplifier (LNA), 222
Low-pitch CT, 411
Low-rank matrix, MRI reconstruction
brainWeb and NIH, CSMRI
comparison, 498
description, 499
NMSE, 497
SPGL1, 496
k-space measurements, 495
lp-norm minimization, 496
rank minimization, 495
Schatten-p-norm, 496
spatial redundancy, 493
Low-voltage high-frequency transducer, 225
LSI, see Linear spline interpolation (LSI)
LSS, see Light scattering spectroscopy (LSS)
Lumbar spinal fusion, 580
M
Magnetic compatibility
eddy currents generation, 391
magnetic field gradients, 392
PET and SPECT shielding materials, 390
spatial resolution measurements, 392
susceptibility, 391
Magnetic resonance imaging (MRI)
acceleration, CS, see Compressed sensing (CS), MRI acceleration
configurations
precession, 532
resonance, 532
T1rho, see T1rho MRI
MAP inference, see Maximum a posterior (MAP) inference
Markov Random Field (MRF) energy minimization
conditional probability, 640
FFD transformation model, 642
fusion-move approach, 642
Gaussian noise model, 641
MAP inference, algorithms, 641
Mass attenuation coefficient, 343
Maximum a posterior (MAP) inference, 640–642
Maximum intensity projection (MIP) CT, 185, 186
Maximum likelihood (ML)
images, regularization, 437, 438
Kuhn–Tucker conditions, 439
Mean square error (MSE), 648
Medical image registration (MIR)
applications, 620
BM algorithm, 646
closed-form solutions, 634–636
data dissimilarity
feature-based information, 634
intensity inhomogeneities, 633–634
model-based information, 632
mono-modality registration, 633
evaluation
brain image, nonlinear algorithms, 650
gradient-based optimization, 648
misalignments, 648
MSE and PSNR, 648
NIREP, 649
split window validation, 647
TRE and segmentations, 648–649
HAMMER, 647
image representation and imaging terminologies, 621
iterative optimization
line search scheme, 637
search approaches, 639
mathematical formulation, 626
regularization
inverse consistency, 631
invertibility, 631
local rigidity, 632
smoothness, see Smoothness regularization, MIR
volume-preservation (VP), 631–632
x-ray breast images, 627
spatial (geometric) transformations, see Spatial transformations, MIR
Medical image segmentation (MIS)
boundary, region and shape, 663
convex relaxation, 674
corpus callosum (CC), 662
energy function, see Energy functions, MIS
Euler–Lagrange equation, 673, 680–681
expert segmentations, 678
gradient-descent process, 673
graph cuts, 674
multiobjective optimization, 675–676
registration, segmentation, 677
segmentation representation, 668–670
Sobolev-type inner products, 673
training data
ASM and PCA, 671
linear statistics, 671–672, 679–680
Medical imaging
creations inspired, imagination, 21, 22
ICs, 1
innovation drives technology, 10
multiprocessor complexity, 14–16
noninvasive techniques, 21
retinal scanner, 11
telecommunications complements, 7–9
MeTRiCS, see Molecular imaging true color spectroscopic (MeTRiCS)
MIP CT, see Maximum intensity projection (MIP) CT
MIR, see Medical image registration (MIR)
ML, see Maximum likelihood (ML)
Modulation transfer function (MTF)
definition, 97
flat-panel X-ray detectors, 135
simulation and measurement, 97–98
Molecular imaging true color spectroscopic (MeTRiCS)
absorbing species, 80
Beer's law, 68
biomedicine, molecular imaging focuses, 68
calculation, correlation coefficients, 71, 72
conventional OCT imaging, 77–78
detector saturation, 82
DW processing, 75
endogenous and exogenous contrast, 77
fluorescence imaging, 67
images, chick embryos in vivo, 76–78
incoherent fluorescent light, 82
influences absorption spectra, 70
light focus, 68
molar extinction coefficients, 69
oxygen saturation, 71
pfdOCT system, 67
photograph and pfdOCT image, 72, 73
quantitative spectral information, 73
RGB values, 74
sensitivity, 69
SOCT, 68
sodium fluorescein (NaFS), 79–82
supercontinuum laser light source, 67
tomographic images, mouse dorsal skin flap, 78–80
variations across individual vessels, 79
visible region, spectrum, 72
Monochromatic energy image, 161–162
Monolithic crystals, statistical positioning, 316
MRF energy minimization, see Markov Random Field (MRF) energy minimization
MSE, see Mean square error (MSE)
99mTc-MDP bone imaging, 365
MTF, see Modulation transfer function (MTF)
Multi-anode PMT (MA-PMT), 291–292
Multicoil parallel MRI, see Compressed sensing (CS), MRI acceleration
Multimodality imaging systems
alpha blending, PET/CT images, 363
CT attenuation correction, 363–364
CT-based co-registration, 361–362
image co-registration, 362
image display, 363
magnetic compatibility, 390–393
MR attenuation correction, 394
nuclear medicine images, 360
PET/CT and SPECT/CT, clinical applications
calcium scoring, 365
18F–fluorodeoxyglucose (FDG) imaging, 364
111In-labeled imaging, 365
99 mTc-MDP bone imaging, 365
myocardial perfusion imaging, 365
PET/MR and SPECT/MR requirements, 367
PET/MR system, 393
problems
nuclear medicine, 366
soft-tissue contrast production, 367
time scales, 366
x-ray CT scans, artifacts, 366
radionuclide transmission imaging, 361
RGB color channels, 363
“unclear” medicine, 361
whole-body PET scan, 360
Multi-pixel Geiger-mode APD, 386–388
Multiple crystal-photodetector layers, 314
Multislice scanners, 336
Myocardial perfusion imaging, 365
N
National Institute of Health (NIH), 496–498
NCG method, see Nonlinear conjugate gradient (NCG) method
NEC, see Noise equivalent count (NEC)
NECR, see Noise equivalent count rate (NECR)
NIH, see National Institute of Health (NIH)
NIREP, see Nonrigid Image Registration Evaluation Project (NIREP)
NLS, see Nonlinear least squares (NLS)
NMSE, see Normalized mean squared error (NMSE)
NNSENSE, see Norm-regularized SENSE reconstruction (NNSENSE)
Noise equivalent count (NEC), 303
Noise equivalent count rate (NECR), 289
Non-deterministic polynomial (NP) hard problem, 487–488, 495, 496
Nonlinear conjugate gradient (NCG) method, 638
Nonlinear least squares (NLS), 474–475
Nonlinear registration algorithms, 351
Nonrigid Image Registration Evaluation Project (NIREP), 649
Non-separable quadratic surrogate (SQS) function, 447–448
Non-SQS function, see Non-separable quadratic surrogate (SQS) function
Normal end-expiration breath-hold protocol, 407–408
Normalized mean squared error (NMSE)
CaLM MRI reconstruction, 521
CS SENSE reconstruction, 519, 520
GRAPPA and l1SPIRiT
reconstruction, 519
iSENSE CS vs. iSENSE NN, 508–509
proposed and CS reconstruction, 497
reconstruction accuracy, Cartesian undersampling, 492–493
Norm-regularized SENSE reconstruction (NNSENSE)
brain images, iSENSE NN, 509–510
description, 501
GRAPPA, 507
reconstruction accuracy, 508–509
NP hard problem, see Non-deterministic polynomial (NP) hard problem
Numerical interpolation method
linear spline interpolation, 39–40
spectral signal, 39
O
OA, see Osteoarthritis (OA)
OCT, see Optical coherence tomography (OCT)
One-pass list-mode expectation-maximization (OPL-EM) algorithm, 419
OPL-EM algorithm, see One-pass list-mode expectation-maximization (OPL-EM) algorithm
Optical coherence tomography (OCT)
application, 83
chromatic dispersion and compensation, 34–36
chromophores, 83
concept, 25
DW signal processing method, see Dual window (DW) processing method
FD-OCT, see Fourier domain OCT (FD-OCT)
FD-OCT system, schematic, 54
fLCI, see Fourier domain low coherence interferometry (fLCI)
full-range Fourier domain, 34, 35
fundamental imaging modes, 24
GPGPU, 25
high-frequency oscillations, 82
imaging
capabilities, 47
instruments measure structure, 54
tools, 83
interferometry scheme, 53
low coherence interferometry (LCI), 53
and MeTRiCS, see Molecular imaging true color spectroscopic (MeTRiCS)
multidimensional imaging
2D cross-sectional imaging, realtime, 43–45
3D (4D) imaging, real-time, 43, 46, 47
parameters,3D biomedical imaging modalities, 24, 25
phantom, 82
submicronlevel resolutions, 25
Optimization algorithms
gradient-based algorithms, 439–441
Kuhn–Tucker conditions, 439
transfer methods
Ordered-subset EM (OSEM) algorithm
augmented cost function, 450–451
convergence rates, 450
convergent OSEM (COSEM), 451–452
EM surrogate function, 451
sub-objective function, 449–450
Ordered subset-expectation-maximization (OS-EM) technique, 419–420
OSEM algorithm, see Ordered-subset EM (OSEM) algorithm
P
Parallel-beam projections, 193–195
Parallel frequency domain OCT (pfdOCT), 67, 69, 72–74
Parkinson's disease (PD), 583, 584, 610–612
Patient-specific attenuation, 353
PCA, see Principal component analysis (PCA)
PCC-SL, see Phase-cycled composition spin-lock (PCC-SL)
PCfdr algorithm
conditional variables, 605–606
error control criteria and results, multiple testing, 606–607
group-level model, 609
mixed-effect model, 610
partial correlation coefficient, 608
pseudo-code, 605
sample size approaches, 607–608
PCG, see Preconditioned conjugate gradient (PCG)
PD, see Parkinson's disease (PD)
PDD, see Primary diffusion direction (PDD)
PDM, see Point distribution model (PDM)
Peak-Signal-to-Noise Ratio (PSNR), 648
Penalized-likelihood (PL), 438–439, 445, 447
Penn State ultrasound imaging system
FPGA chip, 229
mode set, 229
transmit signal generator, 230
PET, see Positron emission tomography (PET)
PET and SPECT imaging
radiation detectors, see Radiation detectors
scintillation detectors, 367–369
PET/CT, see Thoracic PET/CT
PET detectors
advanced
direct conversion detectors, 297–298
photodetectors, 297
scintillator configurations, alternative photodetector, 293–296
semiconductor photodetectors, 292–293
collinear emission, 274
field-of-view (FOV), 274
high-performance
DoI design, see Depth-of-interaction (DoI)
fast and high-Z scintillation materials, 320–321
high photon detection sensitivity, 303–307
improved spatial resolution, see Improved spatial resolution
SiPM detectors, see SiPM devices
ToF PET, see Time-of-flight PET (ToF PET)
noise
detection efficiency, 284
multiple coincidence, 285
prompt coincidences, 285
sources, 284
photodetectors, 297
scintillators, see Scintillators
spatial resolution issues
light sharingtechnique, 282
line-of-response (LOR), 281–282
noncollinearity and positron range, 280–281
pixilated detector solutions, 282
radial elongation, 282
technological contribution, 281–284
pfdOCT, see Parallel frequency domain OCT (pfdOCT)
Phase-cycled composition spin-lock (PCC-SL), 578
Phased-array focusing, 259
Phase locked loop (PPL), 224
Phoswich design, 315
Photodetectors, see Photomultiplier tube (PMT)
Photodiodes
avalanche, see Avalanche photodiode (APD)
capacitance, 381
composition, 380
leakage current, 381
multi-pixel Geiger-mode avalanche, 386–388
PMTs and, 380
radiation detectors, 381
thermal ionizations, 381
Photomultiplier tube (PMT)
capacitor, 372
collector anode, 370
dynode, 370
electron multiplication stage, 369–370
magnetic field effect, 372
multiplier tube efficiency, 370
photomultiplier tube, 369
primary photons, transition, 372
quantum efficiency (Q.E.), 369
secondary electrons, 372
voltages configuration, 370–371
PHS, see Pulse height spectrum (PHS)
PL, see Penalized-likelihood (PL)
Point distribution model (PDM), 671, 675
Point spread function (PSF), 307
Portable high-frequency ultrasound imaging system
B-mode
imaging system hardware, 221–223
ultrasound imaging, see Ultrasound imaging
challenges
hardware specifications, 226
low-voltage high-frequency transducer, 225
CMUT, 219
conventional system's size, 218
Penn State
A/D converters, see Analog-to-digital converters (ADCs)
SRAM, see SRAM
transmitter, see Transmitter
silicon surface detection, 218
SonicWindow, 219
ultrasound ASIC chips, 218–219
Position-sensitive APDs (PSAPDs), 384–385, 395
Position-sensitive PMT (PS-PMT)
PET detector configuration, 292
Positron emission tomography (PET); see also Thoracic PET/CT
detectors, see PET detectors
drug development, 110
micro-CT systems, designs, 111
PET/CT, see Thoracic PET/CT
radioisotope, local activity density, 273
statistical methods, 132
Powell's method, 639
PPL, see Phase locked loop (PPL)
Preamplifier, 222
Preconditioned conjugate gradient (PCG), 440, 452, 453
Primary diffusion direction (PDD)
description, 541
FACT and TEND approaches, 549
probabilities, 552
statistical bootstrap, 553
Principal component analysis (PCA), 671
Probabilistic tractography
Bayesian formulations, 552
diffusion profile, 552
heuristic approaches, 552
limitations, 553
Markov chain Monte Carlo (MCMC), 551
statistical bootstrap, 552–553
Projection-space material decomposition, 158–159
Prototype experimental board, 246
PSAPDs, see Position-sensitive APDs (PSAPDs)
PSF, see Point spread function (PSF)
PSNR, see Peak-Signal-to-Noise Ratio (PSNR)
PS-PMT, see Position-sensitive PMT (PS-PMT)
Pulse-echo experiments
Pitch-Catch mode experimental setup, 248
test results, 249
Pulse height spectrum (PHS), 99, 100
Q
QN methods, see Quasi-Newton (QN) methods
QSCA, see Quadratic surrogate and coordinate ascent (QSCA)
Quadratic surrogate and coordinate ascent (QSCA)
optimal curvature, 447
Poisson log-likelihood function, 446
Quasi-Newton (QN) methods, 638
R
Radial basis functions (RBFs), 624
Radiation detectors
scintillation detectors, 379
solid-state detectors, 388–390
Radiation dose
acquisition protocols, 139
coronal image, 139
maximum intensity projection image, 139, 140
measurement, 139
soft-ware and hardware tools, 138
Radionuclide transmission imaging, 361
Radiotracers, 349
Random coincidences (R), see Random counts
Random counts
coincidence events, detection, 289
ENF and, 289
narrow timing window, 288
NEC curve, 290
timing resolution, 289
RBFs, see Radial basis functions (RBFs)
Receiver
amplified signals, VGA functioning, 234
circuit design details, 231–235
dynamic range, 231
effective bit resolution, 235
Gilbert-type four-quadrant multiplier, 233, 234
protection devices, 225
required specifications, 230–231
transceiver chip gain and dynamic range requirements, 232
Regions-of-interests (ROIs)
brain, 611
functional connectivity, 596
perturbation, 614
seed-based method, 597
Respiratory artifacts, thoracic PET/CT
attenuation correction, 404–405
breathing instruction, 407–410
deconvolution techniques, 421–422
description, 403
gamma rays, 404
gated 4D PET/CT, see Gated four-dimensional (4D) PET/CT
lung carcinoma lesions, 405, 406
ROI, see Regions-of-interests (ROIs)
Rotary echo spin-lock (SL) pulse
B0 inhomogeneities, magnetization evolution, 575
longitudinal magnetization, 576
T1rho-weighted contrast, 575
R1rho relaxation rate, 583
S
SAR, see Specific absorption rate (SAR)
Scatter coincidences (S), see Scatter counts
Scatter counts
Compton scattering, 286
energy spectrum generation, 287
finite energy window, 285
full energy peak, 287
intercrystal scatter, 286
in PET, 303
representation, 285
scatter corrections, 288
scatter fraction (SF), 287
Scintillators
configurations, alternative photodetector
APDs matrices, 294
depth-of-interaction, estimation, 296
DOI error, 296
drawback, 295
monolithic crystals, 295
one-to-one coupling, 294
small animal, 296
spatial resolution issue, 295
detectors
advantages, 379
Compton effect, 369
high-energy gamma ray, 368
high light output, 368
inorganic crystalline material, 368
PET scintillators, 368
properties, 368
radionuclide imaging, 368
effective atomic number, 277
gamma-ray detectors, 277
inorganic, characteristics, 278
light yield, 277
photodetector coupling, 277
SDF, see Signed distance function (SDF)
SD-OCT, see Spectrometer-based spectral domain OCT (SD-OCT)
Segmentation representation, MIS
explicit contour model, 668
fully automatic segmentation, 680
T-snakes and T-surfaces, 669
vessels, 670
Semiconductor detectors, medical imaging
amorphous selenium detectors, 90
indirect and direct conversion detectors, 91–94
intrinsic energy resolution, 105
signal transport processes
anode signals, 97
direct conversion detector, 96–97
indirect conversion detector, 94, 96
spatial resolution
pixel pitch, 97
SPECT, CT and radiography, 89, 90
spectral resolution
counting direct conversion detectors, 103–105
DRF, see Detector response function (DRF)
integrating indirect conversion detectors, 102–103
nuclear physics and medical imaging, 99
tungsten tube spectra, 99, 100
Semiconductor photodetectors
APDs, 292
characteristics comparison, 294
continuous slab solution, 292
Sensitivity encoding (SENSE), MRI reconstruction
brain data, 506
coil sensitivity parameters, 502
ex vivo and in vivo images, rat's spinal cord, 506
iSENSE method, see iSENSE method
JSENSE, see Joint image reconstruction and sensitivity estimation in SENSE (JSENSE)
least squares optimization, 500–501
polynomial coefficients, 502
rank-deficient matrices, 503–504
sensitivity maps and decay of singular values, 503
SparSENSE and NNSENSE, 501
sum-of-squares method, 506–507
TV regularization, 502
Separable quadratic surrogate (SQS) algorithm, 448–449
Shallow breathing CT, 407
Short-time Fourier transform (STFT), 55–57
Signal-to-noise ratio (SNR)
cross-contamination, 165
iodine, 163
material density image, 164
significant improvement, 164–165
two-pass algorithm, 165, 167–168
Signed distance function (SDF), 669, 670, 679
Silicon photomultiplier (SiPM), 292–293
Simultaneous acquisition of spatial harmonics (SMASH), 499
Single-crystal layer/dual-ended photodetectors, 314–315
Single photon emission computed tomography (SPECT), 110, 111, 128, 132, 137, 208–210
Singular value decomposition (SVD), 493, 498
SiP, see System in package (SiP)
SiPM, see Silicon photomultiplier (SiPM)
SiPM devices
digital, see Digital SiPM
Small 2D array
catheter-based CMUT system, 264
CMUT catheters, growth rate, 264
microlinear catheter, 265
real-time volumetric imaging, 265
ring array designs, 264
SMASH, see Simultaneous acquisition of spatial harmonics (SMASH)
Smoothness regularization, MIR
diffusion and curvature, 628
elastic regularization, 629–630
spatial transformation, 627
viscous-fluid-based model, 630
SNR, see Signal-to-noise ratio (SNR)
SoC, see System on a chip (SoC)
SOCT, see Spectroscopic OCT (SOCT)
Software registration, 349–351
Solid-state detectors
Si(Li) and Ge(Li), 388
Sparse modeling, MRI reconstruction
Cartesian sampling
Haar wavelets, 492
rat (spine), brain and phantom, 492
reconstructed images, 493, 494
lp-norm minimization, 489
orthogonal and redundant wavelets, 491
total variation (TV) minimization, 490
wavelet transform coefficients, 489
Spatial transformations, MIR
description, 623
linear and nonlinear models
DFT and DCT, 625
RBFs, 624
terminologies, 623
Specific absorption rate (SAR)
compressed sensing, 579
definition, 578
partial k-space acquisition approach, 579
reduction, spin-lock frequency, 579
spine and cartilage T1rho imaging, 579–580
spin-lock times (TSLs), 579
U.S. FDA, 578
SPECT, see Single photon emission computed tomography (SPECT)
Spectral projected gradient L1 (SPGL1), 496, 498
Spectrometer-based spectral domain OCT (SD-OCT), 29
Spectroscopic OCT (SOCT), 54–56, 68
SPGL1, see Spectral projected gradient L1 (SPGL1)
Spin-lock radiofrequency (RF) pulse
B1 and B0 insensitive composite spin-lock pulse, 572–573, 576–577
Dixon's composite spin-lock pulse and phase cycling, 577–578
magnetization
preparations, 568
MRI hardware, 572
nominal and true spin-lock pulse strength, 572, 573
normal spin-lock pulse, 568, 574
T1rho-weighted contrast, 569–570
SQS algorithm, see Separable quadratic surrogate (SQS) algorithm
SRAM
capacity, 241
clock modulation, simulation results, 243
functional block diagram, 242
measured outputs, 243
on-chip memory, 241
required specifications, 241
SS-OCT, see Swept-laser-based swept-source OCT (SS-OCT)
SSV technique, see Systematic size variation (SSV) technique
STFT, see Short-time Fourier transform (STFT)
Streamline tractography
FACT and TEND approaches, 549
limitations, 550
multitensor models and HARDI, 550
Runge–Kutta method and PDD, 548–549
SVD, see Singular value decomposition (SVD)
Swept-laser-based swept-source OCT (SS-OCT), 29
Systematic size variation (SSV) technique, 237
System in package (SiP), 19–21
System on a chip (SoC)
implementing digital circuits, 20
programmable processors, 21
single-chip calculator, 19
T
Target registration error (TRE), 648
TD-OCT, see Timedomain OCT (TD-OCT)
Telecommunications complements medical imaging, 7–9
TEND approach, see Tensor deflection (TEND) approach
Tensor deflection (TEND) approach, 549
TFT, see Thin-film transistor (TFT)
Theory of compressed sensing (CS), 486–488
Thin-film transistor (TFT), 118, 122
Thin-film ultrasound transducer array
CMOS transceiver chip specifications, 247
PZT layers, 246
SEM image, 247
Sol-gel and multilayer dry-etching, 247
Thin-plate-spline (TPS) model, 624, 625, 628–629, 634, 646, 647
Thoracic PET/CT
breathing instruction-based methods, 407–410
gated 4D, see Gated four-dimensional (4D) PET/CT
low-pitch CT, 411
Timedomain OCT (TD-OCT), 27–28
Time-of-flight PET (ToF PET)
Cherenkov radiators, 318
conventional PET, 318
SNR improvements, 318
ToF PET, see Time-of-flight PET (ToF PET)
Tomographic image reconstruction, 131–132
TPS model, see Thin-plate-spline (TPS) model
Tracer kinetic analysis, PET and SPECT
compartmental model
receptor-ligand binding, 468–471
noncompartmental model
description, 471
spectral analysis, 473
parameter estimation and parametric images
procedures, 464
standard kinetic parameters, 464
Transmitter
beam focusing, programmable delays, 245
CMOS technology, 244
delay time, 244
dynamic focusing and steering delay, 244
focused signal, expression, 243
transmit pulses, 245
TRE, see Target registration error (TRE)
T1rho MRI
applications
BDL and carbon tetrachloride intoxication, 584–585
BT4C glioma animal model, 586
cerebral ischemia, T1rho dispersion, 584
intervertebral disc and articular cartilage, 580–583
murine radiation–induced fibrosarcoma model, 586
myocardial infarction (MI), 586
and T2 relaxation times, 581–582
Bloch–McConnell equations, 571
proton exchange, 571
SAR, see Specific absorption rate (SAR)
spin-lock RF pulse, see Spin-lock radiofrequency (RF) pulse
T1 and T2 relaxation time
CT, x-ray and nuclear medicine, 565–566
Larmor frequency, 566
low-frequency motional processes, 567
precession rate, spins, 567
spin–lattice relaxation rate, 566
spin-lock pulse field strength, 566–567
weighted contrast imaging, 570–572
True coincidences (T), 302
U
Ultrasound imaging
axial resolution, 221
reflection, 220
resolution, 220
V
Voltage-sensitive preamplifier(VSA), 372–374
VSA, see Voltage-sensitive preamplifier(VSA)
W
X
X-ray micro-CT systems
components
angular position, 124
constant radiation level, 122
mechanical subsystem, 126
minimal beam collimation, 125
pixel size and geometrical configuration, 122
rotating gantry, 123
scanner design, 123
source output window, 125
data acquisition, see Data acquisition, micro-CT systems
description, 111
evaluation
quality, reconstructed images, 136–138
X-ray flat-panel detector, 133–136
geometrical configuration, 126–128
high-resolution imaging, 141
PET and SPECT images, 110
small-animal
anesthesia equipment/vital sign monitoring devices, 112
configuration, in vitro, 112
fast and sensitive detectors, 113
in vivo requirements demand, 112, 113
image quality and radiation delivery, 111