Here we present a case study that illustrates how to apply clustering and outlier techniques described in this chapter in the real world, using open-source Java frameworks and a well-known image dataset.
We will now introduce two new tools that were used in the experiments for this chapter: SMILE and Elki. SMILE features a Java API that was used to illustrate feature reduction using PCA, Random Projection, and IsoMap. Subsequently, the graphical interface of Elki was used to perform unsupervised learning—specifically, clustering and outlier detection. Elki comes with a rich set of algorithms for cluster analysis and outlier detection including a large number of model evaluators to choose from.
Note
Find out more about SMILE at: http://haifengl.github.io/smile/ and to learn more about Elki, visit http://elki.dbs.ifi.lmu.de/.
Character-recognition is a problem that occurs in many business areas, for example, the translation of medical reports and hospital charts, postal code recognition in the postal service, check deposit service in retail banking, and others. Human handwriting can vary widely among individuals. Here, we are looking exclusively at handwritten digits, 0 to 9. The problem is made interesting due to the verisimilitude within certain sets of digits, such as 1/2/7 and 6/9/0. In our experiments in this chapter we use clustering and outlier analysis using several different algorithms to illustrate the relative strengths and weaknesses of the methods. Given the widespread use of these techniques in data mining applications, our main focus is to gain insights into the data and the algorithms and evaluation measures; we do not apply the models for prediction on test data.
As suggested by the title of the chapter, our experiments aim to demonstrate Unsupervised Learning by ignoring the labels identifying the digits in the dataset. Having learned from the dataset, clustering and outlier analyses can yield invaluable information for describing patterns in the data, and are often used to explore these patterns and inter-relationships in the data, and not just to predict the class of unseen data. In the experiments described here, we are concerned with description and exploration rather than prediction. Labels are used when available by external evaluation measures, as they are in these experiments as well.
This is already done for us. For details on how the data was collected, see: The MNIST database: http://yann.lecun.com/exdb/mnist/.
Each feature in a data point is the greyscale value of one of 784 pixels. Consequently, the type of all features is numeric; there are no categorical types except for the class attribute, which is a numeral in the range 0-9. Moreover, there are no missing data elements in the dataset. Here is a table with some basic statistics for a few pixels. The images are pre-centred in the 28 x 28 box so in most examples, the data along the borders of the box are zeros:
|
Feature |
Average |
Std Dev |
Min |
Max |
|---|---|---|---|---|
|
pixel300 |
94.25883 |
109.117 |
0 |
255 |
|
pixel301 |
72.778 |
103.0266 |
0 |
255 |
|
pixel302 |
49.06167 |
90.68359 |
0 |
255 |
|
pixel303 |
28.0685 |
70.38963 |
0 |
255 |
|
pixel304 |
12.84683 |
49.01016 |
0 |
255 |
|
pixel305 |
4.0885 |
27.21033 |
0 |
255 |
|
pixel306 |
1.147 |
14.44462 |
0 |
254 |
|
pixel307 |
0.201667 |
6.225763 |
0 |
254 |
|
pixel308 |
0 |
0 |
0 |
0 |
|
pixel309 |
0.009167 |
0.710047 |
0 |
55 |
|
pixel310 |
0.102667 |
4.060198 |
0 |
237 |
Table 1: Summary of features from the original dataset before pre-processing
The Mixed National Institute of Standards and Technology (MNIST) dataset is a widely used dataset for evaluating unsupervised learning methods. The MNIST dataset is mainly chosen because the clusters in high dimensional data are not well separated.
The original MNIST dataset had black and white images from NIST. They were normalized to fit in a 20 x 20 pixel box while maintaining the aspect ratio. The images were centered in a 28 x 28 image by computing the center of mass and translating it to position it at the center of the 28 x 28 dimension grid.
Each pixel is in a range from 0 to 255 based on the intensity. The 784 pixel values are flattened out and become a high dimensional feature set for each image. The following figure depicts a sample digit 3 from the data, with mapping to the grid where each pixel has an integer value from 0 to 255.
The experiments described in this section are intended to show the application of unsupervised learning techniques to a well-known dataset. As was done in Chapter 2, Practical Approach to Real-World Supervised Learning with supervised learning techniques, multiple experiments were carried out using several clustering and outlier methods. Results from experiments with and without feature reduction are presented for each of the selected methods followed by an analysis of the results.
Since our focus is on exploring the dataset using various unsupervised techniques and not on the predictive aspect, we are not concerned with train, validation, and test samples here. Instead, we use the entire dataset to train the models to perform clustering analysis.
In the case of outlier detection, we create a reduced sample of only two classes of data, namely, 1 and 7. The choice of a dataset with two similarly shaped digits was made in order to set up a problem space in which the discriminating power of the various anomaly detection techniques would stand out in greater relief.
We demonstrate different feature analysis and dimensionality reduction methods—PCA, Random Projection, and IsoMap—using the Java API of the SMILE machine learning toolkit.

Figure 8: Showing digit 3 with pixel values distributed in a 28 by 28 matrix ranging from 0 to 254.
The code for loading the dataset and reading the values is given here along with inline comments:
//parser to parse the tab delimited file
DelimitedTextParser parser = new DelimitedTextParser();parser.setDelimiter("[ ]+");
//parse the file from the location
parser.parse("mnistData", new File(fileLocation);
//the header data file has column names to map
parser.setColumnNames(true);
//the class attribute or the response variable index
AttributeDataSet dataset = parser.setResponseIndex(new NominalAttribute("class"), 784);
//convert the data into two-dimensional array for using various techniques
double[][] data = dataset.toArray(new double[dataset.size()][]);The following snippet illustrates dimensionality reduction achieved using the API for PCA support:
//perform PCA with double data and using covariance //matrix PCA pca = new PCA(data, true); //set the projection dimension as two (for plotting here) pca.setProjection(2); //get the new projected data in the dimension double[][] y = pca.project(data);

Figure 9: PCA on MNIST – On the left, we see that over 90 percent of variance in data is accounted for by fewer than half the original number of features; on the right, a representation of the data using the first two principal components.
Table 2: Summary of set of 11 random features after PCA
The PCA computation reduces the number of features to 274. In the following table you can see basic statistics for a randomly selected set of features. Feature data has been normalized as part of the PCA:
|
Features |
Average |
Std Dev |
Min |
Max |
|---|---|---|---|---|
|
1 |
0 |
2.982922 |
-35.0821 |
19.73339 |
|
2 |
0 |
2.415088 |
-32.6218 |
31.63361 |
|
3 |
0 |
2.165878 |
-21.4073 |
16.50271 |
|
4 |
0 |
1.78834 |
-27.537 |
31.52653 |
|
5 |
0 |
1.652688 |
-21.4661 |
22.62837 |
|
6 |
0 |
1.231167 |
-15.157 |
10.19708 |
|
7 |
0 |
0.861705 |
-6.04737 |
7.220233 |
|
8 |
0 |
0.631403 |
-6.80167 |
3.633182 |
|
9 |
0 |
0.606252 |
-5.46206 |
4.118598 |
|
10 |
0 |
0.578355 |
-4.21456 |
3.621186 |
|
11 |
0 |
0.528816 |
-3.48564 |
3.896156 |
Table 2: Summary of set of 11 random features after PCA
Here, we illustrate the straightforward usage of the API for performing data transformation using random projection:
//random projection done on the data with projection in //2 dimension RandomProjection rp = new RandomProjection(data.length, 2, false); //get the transformed data for plotting double[][] projectedData = rp.project(data);

Figure 10: PCA and Random projection - representations in two dimensions using Smile API
This code snippet illustrates use of the API for Isomap transformation:
//perform isomap transformation of data, here in 2 //dimensions with k=10 IsoMap isomap = new IsoMap(data, 2, 10); //get the transformed data back double[][] y = isomap.getCoordinates();
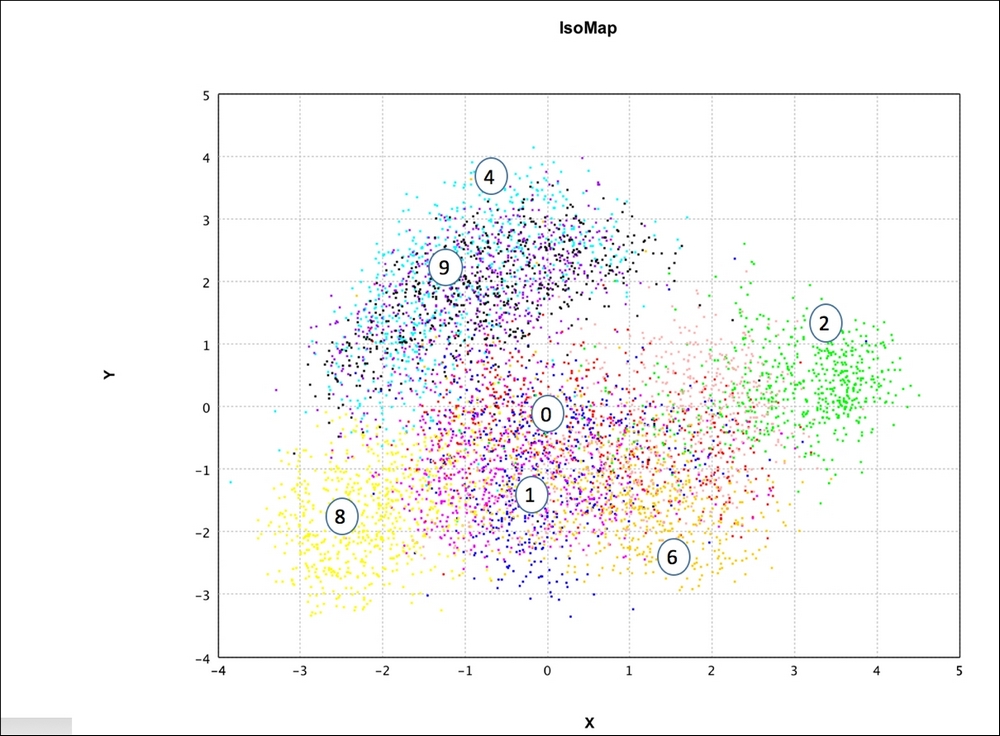
Figure 11: IsoMap – representation in two dimensions with k = 10 using Smile API
We can make the following observations from the results shown in the plots:
- The PCA variance and number of dimensions plot clearly shows that around 100 linearly combined features have a similar representation or variance in the data (> 95%) as that of the 784 original features. This is the key first step in any unsupervised feature reduction analysis.
- Even PCA with two dimensions and not 100 as described previously shows some really good insights in the scatterplot visualization. Clearly, digits 2, 8, and 4 are very well separated from each other and that makes sense as they are written quite distinctly from each other. Digits such as {1,7}, {3,0,5}, and {1,9} in the low dimensional space are either overlapping or tightly clustered. This shows that with just two features it is not possible to discriminate effectively. It also shows that there is overlap in the characteristics or features amongst these classes.
- The next plot comparing PCA with Random Projections, both done in lower dimension of 2, shows that there is much in common between the outputs. Both have similar separation for distinct classes as described in PCA previously. It is interesting to note that PCA does much better in separating digits {8,9,4}, for example, than Random Projections.
- Isomap, the next plot, shows good discrimination, similar to PCA. Subjectively, it seems to be separating the data better than Random Projections. Visually, for instance, {3,0,5} is better separated out in Isomap than PCA.
Two sets of experiments were conducted using the MNIST-6000 dataset. The dataset consists of 6,000 examples, each of which represents a hand-written digit as greyscale values of a 28 x 28 square of pixels.
First, we run some clustering techniques to identify the 10 clusters of digits. For the experiments in this part of the case study, we use the software Elki.
In the first set of experiments, there is no feature-reduction involved. All 28x28 pixels are used. Clustering techniques including k-Means, EM (Diagonal Gaussian Model Factory), DBSCAN, Hierarchical (HDBSCAN Hierarchy Extraction), as well as Affinity Propagation were used. In each case, we use metrics from two internal evaluators: Davies Bouldin and Silhouette, and several external evaluators: Precision, Recall, F1 measure, and Rand Index.

Figure 12: K-Means – using Sum of Squared Errors (SSE) to find optimal k, the number of clusters. An elbow in the curve, which is typically used to pick the optimal k value, is not particularly detectable in the plot.
In the case of k-Means, we did several runs using a range of k values. The plot shows that the Sum of Squared Errors (SSE) metric decreases with k.
The table shows results for k=10 and ranks for each are in parentheses:
|
Algorithm |
Silhouette |
Davies-Bouldin Index |
Precision |
Recall |
F1 |
Rand |
|---|---|---|---|---|---|---|
|
K-Means Lloyd |
+-0.09 0.0737 (1) |
2.8489 (3) |
0.4463 (3) |
0.47843 (3) |
0.4618 (1) |
0.8881 (3) |
|
EM (Diagonal Gaussian Model Factory) |
NaN |
0 (1) |
0.1002 (6) |
1 (1) |
0.1822 (4) |
0.1003 (5) |
|
DBSCAN |
0 (4) |
0 (1) |
0.1003 (5) |
1 (1) |
0.1823 (3) |
0.1003 (5) |
|
Hierarchical (HDBSCAN Hierarchy Extraction) |
+-0.05 0.0435 (3) |
2.7294 |
0.1632 (4) |
0.9151 (2) |
0.2770 (2) |
0.5211 (4) |
|
Hierarchical (Simplified Hierarchy Extraction) |
NaN |
0 (1) |
1 (1) |
0.0017 (5) |
0.0033 (6) |
0.8999 (2) |
|
Affinity Propagation |
+-0.07 0.04690 (2) |
1.7872 (2) |
0.8279 (2) |
0.0281 (4) |
0.0543 (5) |
0.9019 (1) |
Table 3. Evaluation of clustering algorithms for MNIST data
In the second clustering experiment, the dataset was first pre-processed using PCA, and the resulting data with 273 features per example was used with the same algorithms as in the first experiment. The results are shown in the table:
|
Algorithm |
Silhouette |
Davies-Bouldin Index |
Precision |
Recall |
F1 |
Rand |
|---|---|---|---|---|---|---|
|
K-Means Lloyd |
+-0.14 0.0119 |
3.1830 |
0.3456 |
0.4418 |
0.3878 (1) |
0.8601 |
|
EM (Diagonal Gaussian Model Factory) |
+-0.16 -0.0402 |
3.5429 |
0.1808 |
0.3670 |
0.2422 |
0.7697 |
|
DBSCAN |
+-0.13 -0.0351 |
1.3236 |
0.1078 |
0.9395 (1) |
0.1934 |
0.2143 |
|
Hierarchical (HDBSCAN Hierarchy Extraction) |
+-0.05 0.7920 (1) |
0.0968 |
0.1003 |
0.9996 |
0.1823 |
0.1005 |
|
Affinity Propagation |
+-0.09 0.0575 |
1.6296 |
0.6130 (1) |
0.0311 |
0.0592 |
0.9009 (1) |
|
Subspace (DOC) |
+-0.00 0.0 |
0 (1) |
0.1003 |
1 |
0.1823 |
0.1003 |
Table 4. Evaluation of clustering algorithms for MNIST data after PCA
As shown in tables 2.1 and 2.2, different algorithms discussed in the sections on clustering are compared using different evaluation measures.
Generally, comparing different internal and external measures based on technical, domain and business requirements is very important. When labels or outcomes are available in the dataset, using external measures becomes an easier choice. When labeled data is not available, the norm is to use internal measures with some ranking for each and looking at comparative ranking across all measures. The important and often interesting observations are made at this stage:
- Evaluating the performance of k-Means with varying k, (shown in the figure) using a measure such as Sum of Squared Errors, is the basic step to see "optimality" of number of clusters. The figure clearly shows that as k increases the score improves as cluster separation improves.
- When we analyze Table 2.1 where all 784 features were used and all evaluation measures for the different algorithms are shown, some key things stand out:
- k-Means and Affinity Propagation both show a large overlap in the Silhouette index in terms of standard deviation and average, respectively (k-Means +-0.09 0.0737; Affinity Propagation +-0.07 0.04690). Hence it is difficult to analyze them on this metric.
- In the measures such as DB Index (minimal is good), Rand Index (closer to 1 is good), we see that Affinity Propagation and Hierarchical Clustering show very good results.
- In the measures where the labels are taken into account, Hierarchical Clustering, DBSCAN, and EM has either high Precision or high Recall and consequently, the F1 measure is low. k-Means gives the highest F1 measure when precision and recall are taken into consideration.
- In Table 2.2 where the dataset with 273 features—reduced using PCA with 95% variance retained—is run through the same algorithms and evaluated by the same measures, we make the following interesting observations:
By reducing the features there is a negative impact on every measure for certain algorithms; for example, all the measures of k-Means degrade. An algorithm such as Affinity Propagation has a very low impact and in some cases even a positive impact when using reduced features. When compared to the results where all the features were used, AP shows similar Rand Index and F1, better Recall, DB Index and Silhouette measures, and small changes in Precision, demonstrating clear robustness.
Hierarchical Clustering shows similar results as before in terms of better DB index and Rand Index, and scores close to AP in Rand Index.
For the outlier detection techniques, we used a subset of the original dataset containing all examples of digit 1 and an under-sampled subset of digit 7 examples. The idea is that the similarity in shape of the two digits would cause the digit 7 examples to be found to be outliers.
The models used were selected from Angular, Distance-based, clustering, LOF, and One-Class SVM.
The outlier metrics used in the evaluation were ROC AUC, Average Precision, R-Precision, and Maximum F1 measure.
The following table shows the results obtained, with ranks in parentheses:
|
Algorithm |
ROC AUC |
Avg. Precision |
R-Precision |
Maximum F1 |
|---|---|---|---|---|
|
Angular (ABOD) |
0.9515 (3) |
0.1908 (4) |
0.24 (4) |
0.3298 (4) |
|
Distance-based (KNN Outlier) |
0.9863 (1) |
0.4312 (3) |
0.4533 (3) |
0.4545 (3) |
|
Distance Based (Local Isolation Coefficient) |
0.9863 (1) |
0.4312 (3) |
0.4533 (3) |
0.4545 (3) |
|
Clustering (EM Outlier) |
0.5 (5) |
0.97823827 (1) |
0.989 (1) |
0.9945 (1) |
|
LOF |
0.4577 (6) |
0.0499 (6) |
0.08 (6) |
0.0934 (6) |
|
LOF (ALOKI) |
0.5 (5) |
0.0110 (7) |
0.0110 (7) |
0.0218 (7) |
|
LOF (COF) |
0.4577 (6) |
0.0499 (6) |
0.08 (6) |
0.0934 (6) |
|
One-Class SVM (RBF) |
0.9820 (2) |
0.5637 (2) |
0.5333 (2) |
0.5697 (2) |
|
One-Class SVM (Linear) |
0.8298 (4) |
0.1137 (5) |
0.16 (5) |
0.1770 (5) |
Table 5 Evaluation measures of Outlier analysis algorithms
In the same way as we evaluated different clustering methods, we used several observations to compare a number of outlier algorithms. Once again, the right methodology is to judge an algorithm based on ranking across all the metrics and then getting a sense of how it does across the board as compared to other algorithms. The outlier metrics used here are all standard external measures used to compare outlier algorithms:
- It is interesting to see with the right parameters, that is, k=2, EM can find the right distribution and find outliers more efficiently than most. It ranks very high and is first among the important metrics that include Maximum F1, R-Precision, and Avg. Precision.
- 1-Class SVM with non-linear RBF Kernel does consistently well across most measures, that is, ranks second best in ROC area, R-Precision and Avg. Precision, and Maximum F1. The difference between Linear SVM, which ranks about fifth in most rankings and 1-Class SVM, which ranks second shows that the problem is indeed nonlinear in nature. Generally, when the dimensions are high (784), and outliers are nonlinear and rare, 1-Class SVM with kernels do really well.
- Local outlier-based techniques (LOF and its variants) are consistently ranked lower in almost all the measures. This gives the insight that the outlier problem may not be local, but rather global. Distance-based algorithms (KNN and Local Isolation) perform the best in ROC area under the curve and better than local outlier-based, even though using distance-based metrics gives the insight that the problem is indeed global and suited for distance-based measures.
Table 1: Summary of features from the original dataset before pre-processing
