Chapter 1
Emphasizing The Role of Proteins in Construction of the Developmental Genetic Toolkit in Plants
The diversity in land plants due to size, shape, and form is the combined result of different developmental processes for adult plant formation from the zygote and their evolution. The ancestral patterning toolkit designed by Floyd [1] for land plants was constructed by using developmentally important gene families that are responsible for flowering plant growth, patterning, and differentiation. In this chapter we search for the candidate genes, which are important for different stages of embryogenesis of the model plant species Arabidopsis thaliana in light of the dynamical patterning modules concept. This is a novel idea that Newman [2] applied to build a developmental genetic toolkit by using a set of genes that mobilize the physical processes that are important in metazoan development. In this chapter we focus on a small number of gene families, which are related to physical characteristics, such as turgor pressure, asymmetric cell division, asymmetric distribution of cellular components, anisotropic expansion, and dynamics in merstemic cell maintenance and finally establish their evolutionary developmental (evo-devo) roles in land plant development with a functional/biological analysis.
1.1 Introduction
In the evolutionary history of the plant kingdom, the plants evolved through increasing levels of complexity, from a freshwater green alga, through bryophytes, lycopods, ferns, and gymnosperms to the complex angiosperms of today. Between about 480 and 360 million years ago (mya), from a simple plant body consisting of only a few cells, land plants (liverworts, hornworts, mosses, and vascular plants) evolved to an elaborate two-phase lifecycle with complex organs and tissue systems [3]. As mentioned above, the diversity of plant kingdom due to size, shape, and form is the combined result of different developmental processes for adult plant formation from the zygote, which evolved progressively [4].
Identifying the genes, which act in the developmental pathways and consequently in determining how they are modified during evolution, is the focus of the field of evolutionary developmental (evo-devo) biology. The fundamental aspects of the plant body plan have been found to be remarkably consistent within the plant kingdom irrespective of vast diversification [5]. Graham et al. [5] identified nine fundamental body plan features that originated during radiation of algae and were inherited by the plant kingdom. In light of these fundamental features, we analyze the evo-devo roles of embryogenesis in plants. In this chapter we select some candidate genes to construct a genetic toolkit for land plants, and establish their biological/functional significances with an evolutionary analysis.
1.2 Evolutionary Developmental (Evo-Devo) Roles in Embryogenesis of Plants (in Developmental Plant Genetic Toolkit Formation)
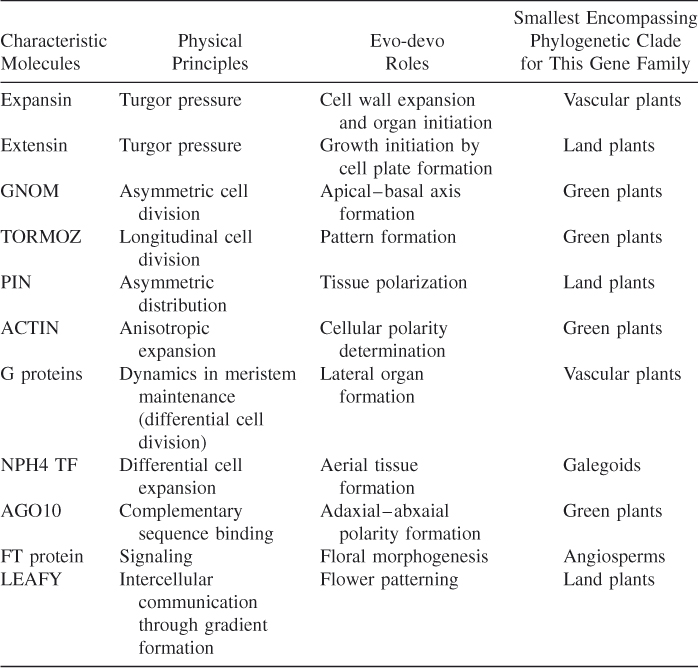
To explore the evo-devo biology of embryogenesis in plants, we select some candidate genes and relate them to their physical properties to depict the developmental genetic toolkit of land plants, following studies by Newman [2]. Table 1.1 shows the candidate genes, their physical principles, and their relevant evo-devo roles.
Table 1.1 Relationship between Candidate Genes and Their Physical Properties and Evolutionary Developmental (Evo-Devo) Roles
| Characteristic Molecules | Physical Principles | Evo-Devo Roles |
| Expansin | Turgor pressure | Cell wall expansion and organ initiation |
| Extensin | Turgor pressure | Growth initiation by cell plate formation |
| GNOM | Asymmetric cell division | Apical–basal axis formation |
| TORMOZ | Longitudinal cell division | Pattern formation |
| PIN | Asymmetric distribution | Tissue polarization |
| ACTIN | Anisotropic expansion | Cellular polarity determination |
| G proteins | Dynamics in meristem maintenance (differential cell division) | Lateral organ formation |
| NPH4 TF | Differential cell expansion | Aerial tissue formation |
| AGO10 | Complementary sequence binding | Adaxial–abxaial polarity formation |
| FT protein | Signaling | Floral morphogenesis |
| LEAFY | Intercellular communication through gradient formation | Flower patterning |
1.3 Phases in Embryogenesis in Arabidopsis Thaliana
In embryogenesis, a multicellular organism forms from a single cell. In Arabidopsis thaliana, embryogenesis is a continuous process that can be divided into three major phases, described as early, mid, and late. In this section, we review all these phases starting from the early phase of embryogenesis.
The early phase consists of pattern formation, morphogenesis, defining the axes of the plant body plan, and forming the organ systems. Embryogenesis involves three basic steps: (1) cell growth, (2) cell differentiation, and (3) morphogenesis. The embryo grows up to certain limit, and then differentiates into cells that differ from their mother cell in structure and function. Thus different morphological structures such as stem, root, or flower are formed, enabling the formation of a total plant.
1.3.1 Cell Growth Phase
The growth in plants is defined as an irreversible increase in cell mass [6]. Since the cell mass value factors both in cell volume and cell number, there are two processes relevant to the plant growth: (1) an irreversible increase in cell size, known as cell elongation, and (2) an increase in cell number, defined as cell division.
1.3.1.1 Cell Elongation Phase
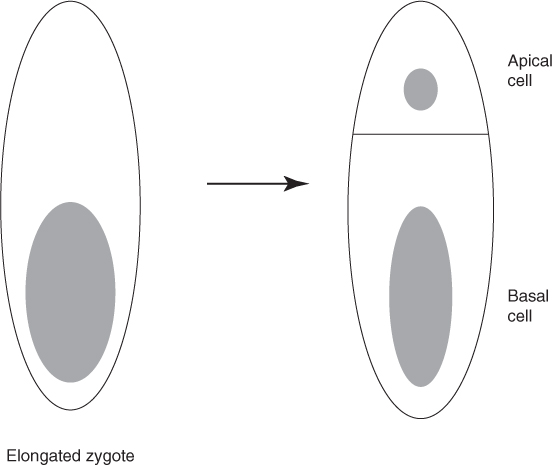
Cell expansion, driven by the turgor pressure, mediates the plant growth. During this process, cells increase manyfold in volume and become highly vacuolated. The cell membrane and the cell wall surround the plant cell, whereas for an animal cell, only cell membrane is present. Since the structure of the cell wall is more rigid than that of the cell membrane, the cell growth mechanism differs in plants and animals. Because of the presence of the rigid cell wall, no cell migration occurs during plant development. During embryogenesis, the zygote elongates 3 times before cell division as a result of the extension of the cells along the sidewalls. Before cell division, the cell nucleus moves to the position of the new cell walls to be formed [7]. Two cells are formed from the zygote following the asymmetric cell division: a small apical cell and a larger basal cell with different cell contents. The dense cytoplasm is present in the apical cell and in most of the basal cell vacuole (see Fig. 1.1).
Figure 1.1 Asymmetric cell division of the zygote.
The steps listed below (1–6) in generation of the embryo proper and the suspensor from the apical and the basal cell, respectively, are shown in Figure 1.2a:
Figure 1.2 Generation of the embryo proper and the suspensor: (a) the steps involved in formation of the embryo proper and the suspensor; (b) the plane of cell division in each step: horizontal —, longitudinal ⊥, and transverse /.
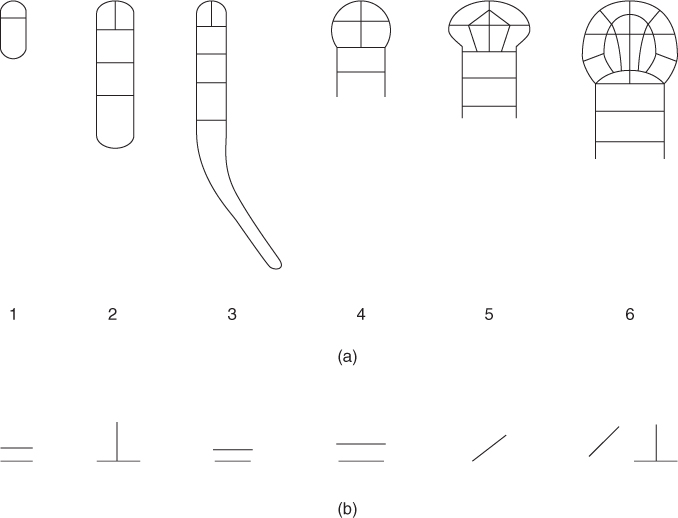
The topmost cell of the suspensor has divided transversely to produce the hypophysis.
The zygote cell initially divides along apical–basal axis. Then the cell growth or expansion occurs. By a few successive cell divisions along different planes, the apical cell and the basal cell, respectively, form the embryo proper and the suspensor. In the embryo proper, eight cells are formed by two vertical and one horizontal cell divisions, whereas the basal cell divides along vertical axis only. The embryo proper will form most of the mature embryo, which are cotyledons, shoot meristem, hypocotyl regions, and part of the radical (or embryogenic) root. From the basal cell, the suspensor and the hypophysis are formed. The hypophysis contributes to the root meristem. Thus, cell growth and expansion in a specific direction and selection of a division plane, all play important roles in plants morphogenesis.
The irreversible increase in cell volume and surface area, as in the cell wall, are determinants of plant cell growth. Both intrinsic and extrinsic factors are required for cell growth. Both light and gravitational forces act as extrinsic factors, whereas the turgor pressure and many other biomolecules are intrinsic factors. Turgor pressure can be generated by water uptake of growing cells in their vacuoles. Since this pressure is homogeneous and multidirectional, the cell wall expands uniformly over the whole cell. Cell wall expansion consists in two processes: (1) stretching of the cell wall resulting from turgor pressure and (2) deposition of new material by the cell membrane in a specific direction. These two processes are related. During cell growth, the cell wall is stretched by turgor pressure because interpolymeric bonds in the wall naturally break and re-form. Thus the polymers forming cell wall, under tension from the turgor pressure, tend to slip past each other irreversibly to enlarge the cell wall. When the cell wall is stretched (normally 10–100-fold), it does not become thinner because new material is deposited to resist the strain of turgor pressure [8]. So the growing cell secretes substances to form a polymeric structure of crystalline cellulose microfibrils, embedded in a hydrophilic matrix, which are composed of hemicelluloses and pectins.
1.3.1.1.1 Role of Expansin and Extensin Proteins in Cell Elongation
Cosgrove [9] proposed the role of protein expansins in cell wall expansion by slippage or rearrangement of matrix polymers. Plant cell wall expansion occurs more rapidly at low pH. This event is known as acid growth. The primary cell wall of plant is composed of xylans (homopolymers of xylose), xyloglucans (heteropolymers whose backbone consists of glucose and sidechains with xylose), and mixed linked glucans (homopolymers of glucose). Xlycan (polysaccharide) molecule are present in the cell wall. They can stick to each other, as well as connect the cellulose microfibrils to each other. Expansins disrupt both types of bonding: between xlycans and between xlycan and cellulose. In presence of turgor pressure, expansins cause the displacement of wall polymers and a slippage occurs at the point of polymer adhesion. In shoot apical meristem, the expansins are expressed, and these proteins are present in primordium forming cells. During leaf initiation, the cell wall is altered by expansins. Thus, in presence of the turgor pressure, the cell wall of meristem expands in a specific direction; For example, it may bulge outward to form primodium. This is the regulatory role of the cell wall protein (expansin) in leaf initiation (plant morphogenesis) [10].
Extensins are a family of hydroxyproline-rich glycoproteins (HRGPs), which are found in the cell walls of higher plants. They play important roles in determining the cell shape and formation of the division plane during cell division. In dicots, extensins have a repeated pentapeptide Ser–(Hyp)4 structure. It has been proposed that a positively charged extensin scaffold reacts with acidic pectin to form extensin pectate. This can act as the template for a newly synthesized cell plate during cell division. Thus extensins play an essential role in growth initiation of plants.
The RSH gene is a lethal mutant in Arabidopsis embryogenesis that encodes extensin AtEXT3, a structural glycoprotein located in the nascent cross wall or “cell wall” and also in mature cell walls. RSH is essential for the correct positioning of the cell plate during cytokinesis in cells of the developing embryo. RSH is detected in the first asymmetric cell division of the zygote [11]. In the RSH mutant, both apical and basal cells are formed, but the position of the plane of the newly synthesized cell wall is different, which may be compared to that of wild types. As a result, this event forms either same sized apical and basal cells or larger apical cells. This type of mutant also affects embryonic development in later stages, such as the globular stage. Cell division occurs during this stage, but the specific plane of division, as shown in Figure 1.2b, is not maintained. Therefore, cell divisions occur randomly. These stop the normal bilateral symmetry of embryogenesis.
The major component of the plant cell wall is polysaccharides (80% present in Arabidopsis). Pectin and hemicellulose are the main polysaccharides. In the mature cell wall, pectin is present in middle lamella, which adheres two adjacent plant cells to each other. Hemicellulose is involved in cell expansion, cell growth, and thus in cell shape formation. Both pectin and hemicellulose are synthesized and modified in Golgi complex. After packing into different vesicles (specific for each component), they are targeted to different domains of the cell wall. This type of physical segregation of components of cell wall is important for the normal growth and the development of plants. Extensin proteins, as mentioned earlier, are modified in cis-Golgi network and through secretary vesicles delivered to trans-Golgi network and ultimately to cell wall. Thus cell growth occurs. This explains the importance of selecting expansin and Extensin proteins in the genetic toolkit for plant cell elongation.
1.3.1.2 Cell Division Phase
After elongation in a specific direction, cell division occurs in a specific plane, so that cell partition occurs along its longest axis. This is observed in the first cell division of the zygote, where apical–basal axis is formed by an asymmetric cell division.
1.3.1.2.1 Role of GNOM for Normal Cell Division
From the very first cell division, Shevell et al. [12] observed that orientation of the plane of cell division, the rate of cell division, and GNOM gene products control the direction and amount of cell expansion. In the GNOM mutant, instead of asymmetric cell division in the zygote, two nearby equal-sized cells are formed. As a result of the distorted orientation of the plane of cell division, an altered octant embryo is formed with twice the number of cells found in the wild type. No cotyledonary primodia forms from apical cell. The root development is inhibited in the GNOM mutant because the basal cell fails to form the hypophysis. Very little elongation occurs in mutant cells with abnormal vacuoles [12]. In dark, wild-type hypocotyls elongate in the longitudinal direction. But for the dark-grown GNOM mutant, expansion occurs in both horizontal and longitudinal directions [12]. Thus, in the GNOM mutant, regulation on the direction of cell elongation is absent. Another important observation is that GNOM mutant cells can be separated easily from one another. This may be due to the inappropriate deposition of pectin or its derivatives in cell wall. From these observations, we can conclude that GNOM is essential for normal cell division, cell expansion, and cell adhesion in Arabidopsis.
ARF (ADP ribosylation factor) proteins are small guanine–nucleotide binding proteins. They are interconverted between two forms, such as GTP-bound ARFs and GDP-bound ARFs, by guanine–nucleotide exchange factors (GEFs) and by GTPase activating proteins (GAPs), respectively [13]. GNOM is a guanine–nucleotide exchange factor of ARF class proteins. It is present in the cytosolic face of endosomes. Generally ARFs are responsible for formation of vesicle coats, which are necessary for the formation of transport vesicles from donor compartments (i.e., vesicle budding) and cargo selection for transmembrane proteins. Here, GNOM proteins are involved in coat recruitment to endosomes for PIN1 protein targeting. PIN1 is a transmembrane protein, which is responsible for auxin hormone transport. GNOM proteins, as a regulator of intracellular vesicle trafficking, controls the rapid cycling of PIN1 proteins between the basal domain of plasma membrane and the endosomal compartments. Thus the embryos lacking the GNOM protein fail to establish the coordinated polar localization of the auxin–efflux regulator PIN1. This explains our selection of GNOM protein for the evolutionary–developmental (evo-devo) role of cell division in the plant genetic toolkit.
1.3.2 Cell Differentiation Phase
The term differentiation with respect to the plant cell refers to the property of plant cells to form quantitatively different specialized cell groups. During this phase, a number of cells that are derived from a single progenitor cell or a group of cells are qualitatively different in their contents and are specialized for different functions [6].
Several different mechanisms are involved in plant cell differentiation:
In the next section, for each of these mechanisms, we evaluate the importance of associated proteins, to form the plant genetic toolkit.
1.3.2.1 Role of PIN for Signaling through the Cell Wall (Associated Determinants)
Cell polarity is an asymmetric distribution of the cellular components with respect to an arbitrary axis inside the cell [14]. An important example of plant polarity is the apical–basal polarity of the PIN family of auxin efflux facilitators, which forms the organization of the entire plant body. In plants, the matured embryo contents a main axis of polarity, with the shoot meristem flanked by the cotyledons (embryonic leaves) at the top end and separated by hypocotyl (embryonic stem) and root from the root meristem at the opposite pole [15]. This is due to tissue polarization (in addition to cell polarization) during plant development, which is necessary for organogenesis. Auxin hormone as a key regulator of tissue polarization controls its own polar transport in response to internal factors (various transcription factors) and external factors (light, gravity). Thus auxin is distributed asymmetrically along matured embryo.
Auxin is a multifunctional phytohormone that controls various developmental processes, such as cytoskeleton organization, intracellular membrane trafficking, cell polarity and morphogenesis, cell division, cell expansion, cell differentiation, organ formation and growth (lateral organ and root hair production), organization of plant architecture (phyllotaxis), and tropic growth [16]. The asymmetric distribution of auxin depends on polar transport of auxin by different transporter proteins such as PIN and AUX1, which facilitate the auxin efflux out of the cells and influx toward the cells, respectively.
In Arabidopsis the PIN (PINFORMED) family consists of eight members, which are distributed asymmetrically on different sides of various cells. According to the chemiosmotic model of auxin biology, the direction of intercellular auxin movement within the fields of cells is controlled by the position of efflux carriers at one side of the transporting cells. Thus auxin mediates the tissue and organ polarity. During the apical–basal axis formation of embryo, the polar localization and coordinated functions of PIN1, PIN2, PIN3, PIN4, and PIN7 are shown in Table 1.2 (see also Fig. 1.3).
Table 1.2 Tabulation Indicating Direction of Auxin Flow and Localization of PIN Proteins on Plasma Membrane during Four States of Embryogenesis
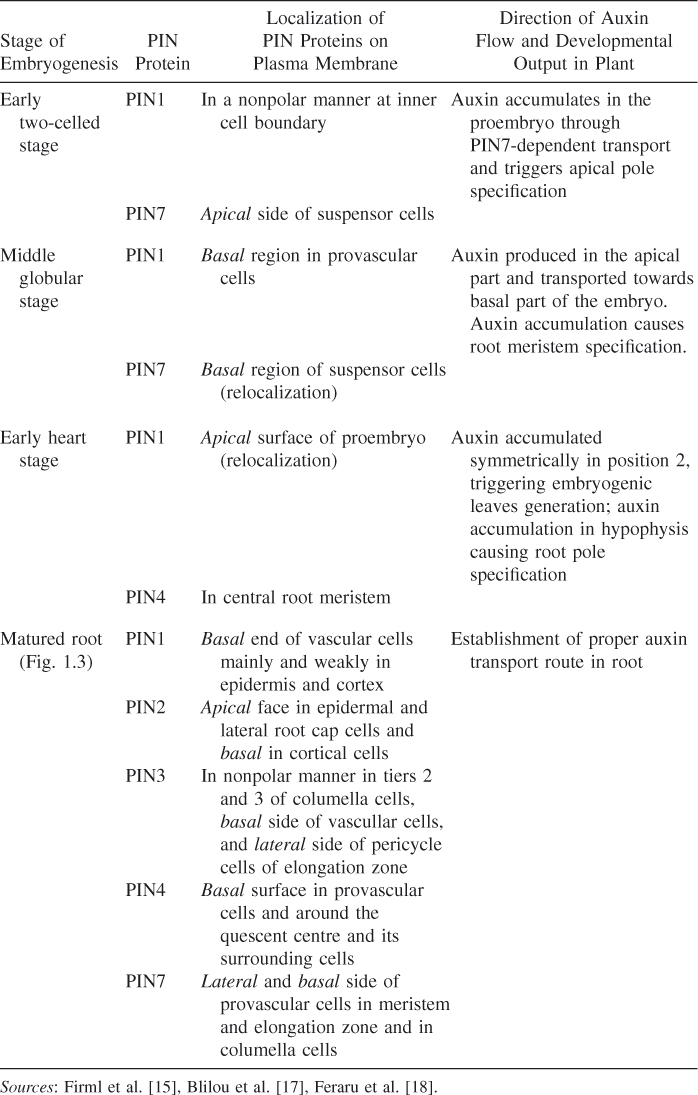
Figure 1.3 Matured root with different developmental regions.
Dhonukshe et al. [19] proposed a two-step mechanism for PIN polarity generation:
In step 2, the clathrin-dependent endocytosis is crucial to establish polarity. The polar PIN localization is dynamic. PIN proteins continuously undergo cycles of clathrin-dependent endocytosis and ARF-GEF-(guanine–nucleotide exchange factors for ADP ribosylation factor GTPases)-dependent recycling such as the GNOM-depedent pathway for PIN1. The internal factor, the Ser/Thr kinase PINOID (PID) [20], plays a central role in the control of apical–basal PIN targeting [15]. The PID phosphorylates Ser337 and/or Thr340 in the central hydrophilic
loop of PIN proteins and PP2A phosphatase antagonizes this action. It has been observed that when PIN is in dephosphorylated form, basal PIN polar targeting occurs, and in phosphorylated state apical trafficking is preferred [21]. Thus the phosphorylaion acts as an internal signal for PIN polarization. PIN polarity forms auxin gradient by controlling the direction of auxin flow, resulting in organ formation in the plants. Furthermore, intact actin cytoskeleton plays an important role in interphase PIN trafficking, whereas this trafficking depends on microtubule during the cell divison phase [22].
1.3.2.2 Role of F-Actin in Cell Polarity (Determinant)
In the plant cell, the cytoskeleton, which is composed of microtubules and actin filaments, mediates cell growth. This cytoskeleton plays an important role in cell division, cell shape determination, cell expansion, and organelle movement. Specifically, three-dimensional (3D) networks of actin filaments are responsible for organelle and vesicle transport. Actin disrupting drugs can alter PIN1 positioning in plasma membrane because actin cytoskeleton controls the cycling of PIN1 between the endosomes and the cell membrane [22]. In columella cells, PIN3 targeting changes from the basal position toward the lateral side of the plasma membrane when the actin cytoskeleton is disrupted. This evokes a bending response in the root [23]. In shoots, actin disruption inhibits polar auxin transport. So, the auxin transport in plant is facilitated by dynamics of actin. It has been observed that intracellular motility of actin filaments will be decreased by stabilized actin in presence of auxin [24]. The bundling of actin causes reduced longitudinal transport of auxin and consecutively hampers root growth in the presence of gravitational force. The opposite result is observed with exogenous auxin addition by restoring normal actin network [25]. Thus it can be concluded that auxin controls its own efflux via actin cytoskeleton.
Phytohormone auxin is the central element for axis formation and pattern formation in plants. Auxin transport inhibitors (ATIs) establish the role of actin cytoskeleton in auxin polar transport. 2,3,5-Triidobenzoic acid (TIBA) and 2-(1-pyrenoyl) benzoic acid (PBA) are two ATIs. They stabilize the actin network by bundling of actin [19]. Thus the actin dynamics is inhibited in the plant. This stabilization hampers actin-dependent trafficking of auxin, causing a disturbance in the endocytosis, vesicle motility, and thus in the auxin transport. As a result, the plant development will be affected, since TIBA and PBA can disturb actin network in plant, by disrupting subcellular trafficking of PIN.
Actin proteins are present in two forms: (1) G-actin or globular subunit and (2) F-actin or filament actin. Both F-actin and actin filaments are polymeric form of monomeric G-actin. Actin binding proteins bundle actin filaments. Bundling of actin is the close alignment of F-actins, in either parallel or antiparallel orientation. These bundles are often aligned with the long axis of the cells. The actin network is composed of complicated configuration of filaments and bundles surrounding nucleus. There are 10 actin genes in the Arabidopsis genome. Among them, eight functional genes can be divided, according to their patterns of expression in plant, into two groups:
These five reproductive genes are expressed predominantly in reproductive organs such as in pollen tubes, but also in some vegetative tissues. Several different cellular processes are controlled by actin proteins:
Arabidopsis-related protein 7 (ARP7) is an essential gene and is required for normal embryogenesis in Arabidopsis. For mutant embryo, the growth is stopped at the heart or torpedo stage [27]. ACT11 is the only Arabidopsis actin gene expressed at significant levels in ovule, embryo, and endosperm. The ACT11 isovariant plays distinct and required roles during Arabidopsis development [28].
1.3.2.3 Role of TORMOZ in Asymmetric Cleavages in Different Cell Planes
During development of the Arabidopsis embryo, the cell fate is determined in a position-dependent manner, since cell migration does not occur in the plant body. To obtain distinct cell types from a single progenitor cell, the asymmetric cell division is a common method. When a plant cell divides, a new cell wall forms between the daughter cells. The positioning of new walls has significant effects on development. By partitioning the cell into unequal parts with differing environments and differing contents, the site of the division plane influences subsequent cell fate. The selection of division plane or orientation of cell division is an important aspect in plant tissue organization and overall organ formation, including morphogenesis. Actin filaments accompanied by microtubules guide the formation of new cell wall during cytokinesis.
During embryogenesis in Arabidopsis, the first few successive cell divisions occur mostly perpendicular to previous divisions as shown in Figure 1.2. The TORMOZ (TOZ) gene encodes a nucleolar protein. The mutation of the TORMOZ (TOZ) gene yields embryos with different cell division planes with abnormal patterns. Longitudinal division planes of the proembryo are changed mainly to transverse divisions and sometimes to oblique divisions. Again, divisions of the suspensor cells, which divide only transversely, appear generally unaffected [29]. Thus, TOZ function is to direct longitudinal cell divisions in the Arabidopsis embryo in the plant developmental gene toolkit.
1.3.2.4 Role of G Protein in Differential Cell Division
In animal the kingdom, heterotrimeric G proteins containing α, β, and γ subunits play important roles in different signaling pathways. Similarly, these types of plants proteins are important for various response reactions for hormones, drought, pathogens, and different developmental processes such as lateral root formation, hypocotyl elongation, and leaf expansion. Cell division is one of several biological processes that can be regulated by G-protein complex. Arabidopsis Gα (GPA1) and Arabidopsis Gβ (AGB1) subunits are more strongly expressed in roots than in shoots in young seedlings. Stem cells of the root apical meristem (RAM) generate different types of cells through cell division, followed by cell elongation. Root growth is a total process, the combined effect of maintenance of cells of root apical meristem (RAM) in the undifferentiated state and lateral root formation. The GTP-bound form of GPA1 accelerates cell division in the RAM. In the apical meristem, the GTP-bound form of the Gα subunit role is a positive modulator for cell proliferation. Lateral root production requires meristem formation by the founder pericycle cells. The β subunit of the Arabidopsis G protein negatively regulates cell division. The Gβγ dimer inhibits cell division in the pericycle founder cells [30]. It was observed that the null alleles of Arabidopsis Gα (gpa1) have a reduced number of lateral root primordia, whereas the null alleles of Arabidopsis Gβ (agb1) have enhanced the cell division in roots, resulting in excessive lateral roots [31]. These results suggest that Arabidopsis heterotrimeric G-protein subunits have differential cell division in roots. Therefore, we select a G-protein subunit to include in the cell division phase of the developmental plant genetic toolkit.
1.3.2.5 Role of NPH4 Transcription Factor in Differential Expansion Rate
The unequal cellular growth in one position of an organ relative to an opposing position is observed in response to the environmental signals This is known as differential growth. Auxin modulates plant growth by regulating the transcription of specific mRNAs that encode proteins necessary for the growth control, such as α expansins.
The NPH4 gene of Arabidopsis is required for auxin-dependent differential growth responses of aerial tissues for both phototropism and gravitropism [32]. NPH4 gene encodes the auxin-regulated transcriptional activator ARF7. Auxin has been proposed to modulate gene expression through modification of ARF activity by auxin-dependent ARF–ARF and ARF–Aux/IAA dimerization. The dissociation–association state of NPH4/ARF7, whether it is present alone or as a complex with MSG2/IAA19 [33], determines the tropic responses of the hypocotyl. Tatematsu et al. [33] proposed a simple regulatory feedback loop for the control of auxin-dependent tropic responses, in which the transcriptional activity of NPH4/ARF7 is controlled by the Aux/IAA repressor protein MSG2/IAA19 via action of IAA-amido synthetases. When auxin accumulates, the ability of NPH4/ARF7 to function as a transcriptional activator is expected to be highest, and cell elongation is stimulated. In response to this event, the gravitropic or phototropic stimuli in hypocotyls two expansin genes, EXPA1 and EXPA8, are upregulated only in the section where the cell elongation occurs [34]. These two tropic stimulus-induced genes, EXPA1 and EXPA8, encode enzymes involved in cell wall extension. This response is essential for the differential growth leading to curvature. It was observed that before the macroscopic curvature EXPA1 and EXPA8 mRNAs expression was increased and cell expansion should be enhanced by the increased expansin delivery at the cell wall in response to auxin. Thus EXPA1- and EXPA8-encoded proteins are directly involved in differential growth response through the activity of NPH4 transcription factor in the developmental gene toolkit in plants.
1.3.2.6 Role of ZWILLE as a Micro-RNA Regulation
Posttranscriptional gene silencing is another method for plant cell differentiation. The small noncoding RNAs play an important role in the gene regulation in association with a unique class of proteins called argonautes [35]. When they are bound with small regulatory RNAs such as short interfering RNAs (siRNAs), or micro-RNAs (miRNAs), argonaute proteins can control protein synthesis by affecting messenger RNA stability.
Arabidopsis contains 10 Argonaute (AGO) proteins. AGO10 is a member of the AGO family. This protein is required to establish the central–peripheral organization of the embryo apex. An empty apex, or a pinlike structure or the solitary leaf in place of the apex was observed in the mutants of AGO10 gene, known as pinhead (pnh) or zwille (zll). AGO10 plays another role in the formation of leaf adaxial–abaxial polarity to specify the leaf adaxial identity in plants [36]. AGO10 does both SAM maintenance and formation of leaf polarity by repressing miR165/166. Micro-RNAs (miRNAs) are ∼21-nucleotide noncoding RNAs. Here two miRNAs, miR165 and miR166, differ in only one nucleotide in their mature RNA sequences. Because PHB, PHV, and REV have a complementary sequence of miR165 and miR166, so they can act as target sites for the cleavage activity of these two miRNAs cleavage activity. In earlier experiments it was shown that abnormally increased miR165/166 levels could result in a severe reduction of HD-ZIP III transcripts such as REV,PHB. The genes, which belong to HD-ZIP III transcription factor family such as PHABULOSA (PHB), PHAVULOTA (PHV), and REVOLUTA (REV), control vascular bundle development and adaxial–abaxial axis formation in leaves. Therefore the plant miRNAs (miR165/166) control the developmental pattern by downregulating important developmental transcription factors in the developmental genetic toolkit.
1.3.3 Plant Morphogenesis Phase
Plant morphogenesis is the development of plant form or shape and structure by coordinated cell division and growth. It is one of the three fundamental aspects of developmental biology along with the control of cell growth and cellular differentiation. The process controls the organized spatial distribution of cells during the embryonic development of an organism. Morphogenesis may be concerned with the whole plant, with a plant part, or with the subcomponents of a structure.
Since the cardiovascular system is absent in plants, plants have evolved specialized transport pathways to distribute signals and nutrients for coordination among different organs of the whole plant as well as different parts of an organ. There are two transport mechanisms: vascular system and intercellular transport pathways for plants. Two vascular networks, the phloem and the xylem, are primarily responsible for longdistance transport. The plasmodesmata, which form a meshwork of plant-specific cytoplasmic tunnels, play major role for intercellular transport.
Several different molecules are related to plant morphogenesis:
1.3.3.1 Role of FT Protein in Longrange Signaling through Low-Molecular-Weight Substances
FLOWERING LOCUS T (FT) is a small globular protein of 20 kDa that serves as a long-range developmental signal in the plants. This mobile signal FT is expressed in the phloem tissues of cotyledons and leaves. It travels from the leaves to the shoot apex, through phloem companion cells, and triggers floral morphogenesis. The long-distance signal, called florigen, initiates flower formation that induces flowering in response to daylength. FT protein in Arabidopsis is an important part of florigen. It has been shown that full FT action in the leaf is dependent on the flowering time gene FD. Environmental factors such as temperature and light intensity, can change the timing of the transition to flowering not only by modifying FT transcript levels but also by modifying FT activity [37]. CONSTANS(CO) gene mediates transcriptional activation of FT under inductive daylength conditions. This gene itself regulates by a complex interplay of signals in the photoperiod pathway.
There are two types of mechanisms for regulation of FT gene expression:
The FT protein acts in the shoot apex to induce target meristem identity genes such as APETALA1 (AP1) and initiates floral morphogenesis [39]. Therefore FT and FD together act redundantly with the floral integrator LEAFY (LFY) to activate AP1 transcription, because a plant containing mutations in both FT and LFY completely lacks floral structures and AP1 expression [37].
1.3.3.2 Role of LEAFY in Shortrange Signaling via Transcription Factor Transfer
For intercellular communication, plants have evolved cytoplasmic bridges, called plasmodesmata, which act as a link of the fluid cytoplasm between adjacent cells. Many transcription factors can move between cells through plasmodasmata. Not all transcription factors move freely in the plant, because there are active mechanisms that regulate transcription factor movement [40]. Shoot apical meristem (SAM) consists of three tissue layers: the outermost layer (L1), the subsurface layer (L2), and the inner layer (L3). Within floral meristems LEAFY is expressed as mRNA in cells of L1 layer. Previously, LEAFY protein was found in a gradient that extended into several interior cell layers [47]. Not only LEAFY can undergo cell-to-cell transport into the underlying L2 and L3 layers, but it also retains its biological activity after transport as a DNA binding transcription factor [41].
The plant-specific transcription factor LEAFY (LFY) has central, evolutionarily conserved roles, both in the formation of the first flower during the meristem identity (MI) transition from vegetative growth and later in flower patterning in angiosperms probably since the origin of flowering plants [42, 43]. LEAFY (LFY) is found in all land plants, which evolved during the past 400 million years, including both flowering and nonflowering plants [44]. But their role in nonflowering plants is not well understood. It has been proposed that LFY homologs have an ancestral role in regulating cell division and arrangement [43]. In flowering plants LFY protein provides only a redundant mechanism to ensure complete conversion of a meristem into a flower by movement of the protein to adjacent cells. During the floral transition, AP1 expression is directly activated by LFY and by a complex consisting of FT and FD [45]. AP1 binds to promoter and regulates the expression of flowering time genes SVP, SOC1, and AGL24. This establishes the significance of LEAFY proteins in the role of short-range signaling via transcription factor transfer in the plant developmental genetic toolkit.
1.4 Analysis
We analyze the candidate genes of the developmental genetic toolkit by using PLAZA 2.0, which integrates structural and functional annotation of 23 plants: 11 dicots, 5 monocots, 2 (club)mosses, and 5 algae. PLAZA is an access point for plant comparative genomics centralizing genomic data produced by different genome sequencing initiatives. It integrates plant sequence data and comparative genomic methods and provides an online platform to perform evolutionary analyses and data mining within the green plant lineage (viridiplantae).
The explicit phylogenetic framework existing in PLAZA framework depicts the early key events and evolution of land plants. Figure 1.4 exhibits a morphological evolution of land plants, consecutively descending from green plants, land plants, vascular plants (monocots), angiosperms, eudicots, and rosids to fabids. For analyzing our embryogenesis toolkit on land plants, we follow another simplified version of phylogenetic species tree, as illustrated in Figure 1.5.
Figure 1.4 Phylogenetic relationships among different plants as inferred from state-of-the-art molecular and morphological phylogenetic analysis (PLAZA, web page).

Figure 1.5 Phylogenetic tree of the evolution of land plants.
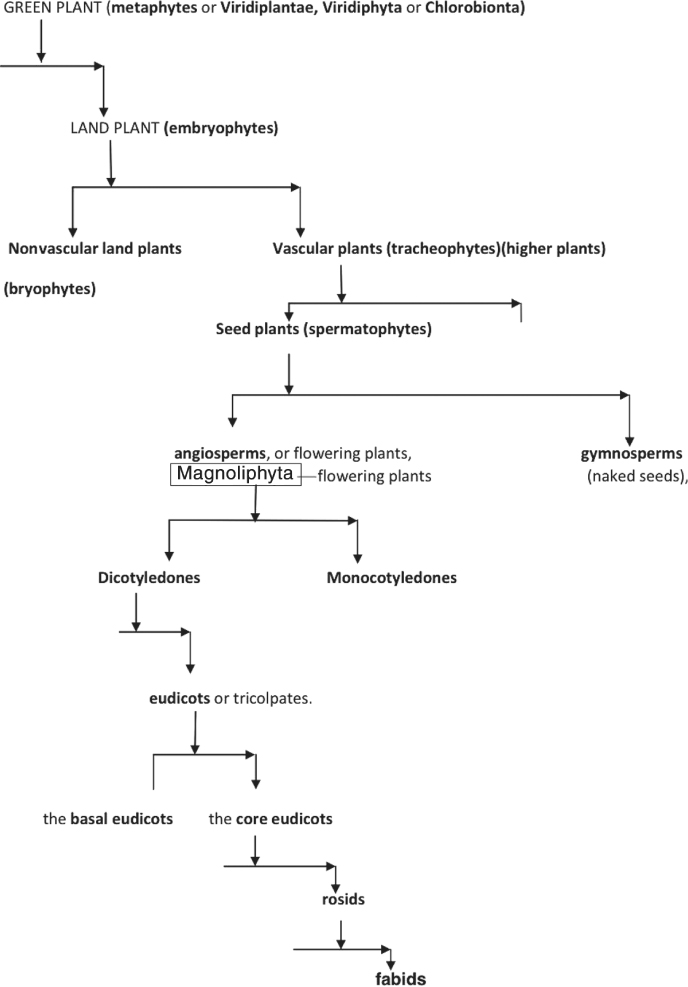
Historically the “plant” phylum implies an association with certain traits, such as multicellularity, cellulose, and photosynthesis. In the evolutionary phylogenetics, green plants are also known as viridiplantae, viridiphyta, or chlorobionta [48]. This clade also includes the land plants plus Charophyta (i.e., stoneworts), and Chlorophyta (i.e., other green algae such as sea lettuce). The Viridiplantae clade encompasses a group of organisms that possess chlorophyll a, b, that have plastids that are bound by only two membranes and are capable of storing starch, and have cellulose in their cell walls. The multicellular land plants, called embryophytes, include the vascular plants, such as plants with full systems of leaves, stems, and roots. Consequently, early seed plants are referred to as gymnosperms (naked seeds) and angiosperms, representing the flowering plants.
Descending further in the evolutionary history, the dicotyledons, also known as dicots, are a group of flowering plants whose seed typically has two embryonic leaves or cotyledons. There are around 199,350 species within this group [49]. Flowering plants that are not dicotyledons are monocotyledons, typically having one embryonic leaf. Descending from this clade, the vast majority of “dicots” do form a monophyletic group called the eudicots or tricolpates. The term eudicots has been widely adopted to refer to one of the two largest clades of angiosperms (constituting >70% of all angiosperms); monocots constitute the other. Eudicots can be divided into two groups: basal eudicots and the core eudicots [49]. The term basal eudicots is an informal name for a paraphyletic group. The core eudicots are a monophyletic group. Within the core eudicots, the largest groups are the rosids, which are a large clade of flowering plants, containing about 70,000 species, [50] more than a quarter of all angiosperms [51]. This group is divided into 16–20 orders, depending on circumscription and classification. These orders, in turn, together comprise about 140 families [52]. The rosids and the asterids are by far the largest clades in the eudicots.
Considering this pattern of phylogenetic history as the baseline for the genetic systems evolution among plants, we further synthesize the phylogenetic relationships of the functions of our chosen developmental genes and their roles in hypothesizing the ancestral land plant developmental toolkit. Figures 1.6–1.16 illustrate the cladograms, which depicts relationships of our chosen orthologous developmental genes in land plants inferred from PLAZA 2.0 [54]. In each figure, the known gene names are labeled next to the lineages on the right side. The labels near each event indicate the clade for descendant lineages mapped onto the simple land plant phylogeny. The labels next to the branches indicate the branch length of the specific plant clade.
Figure 1.6 Phylogenetic and functional evolutionary history of ATEXP1 genes.
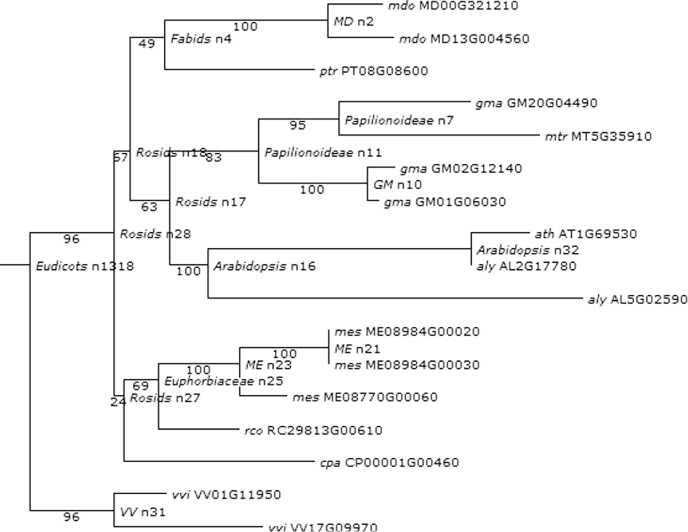
Figure 1.7 Phylogenetic analysis of extensin genes in PHYTOZOME.
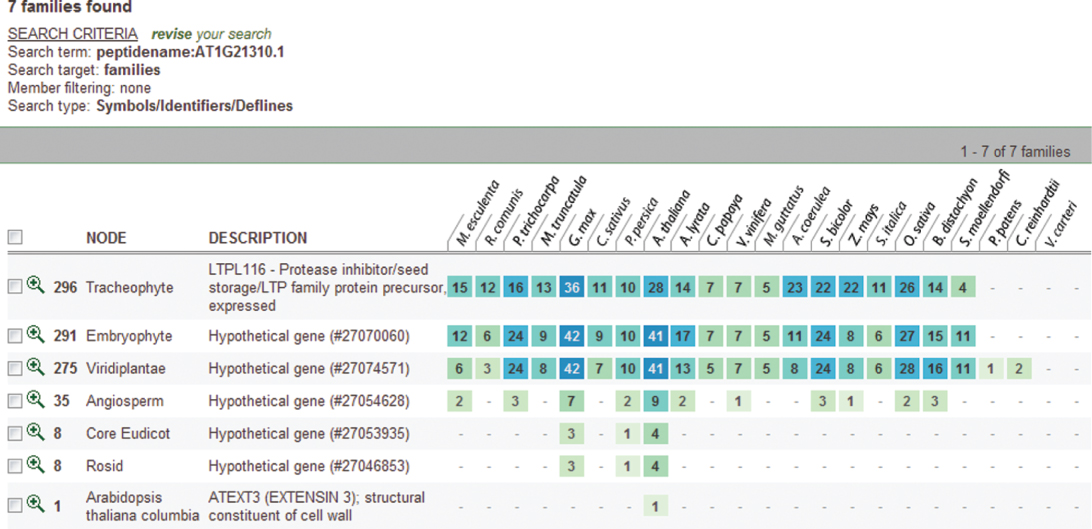
Figure 1.8 Phylogenetic and functional evolutionary history of GNOM genes.
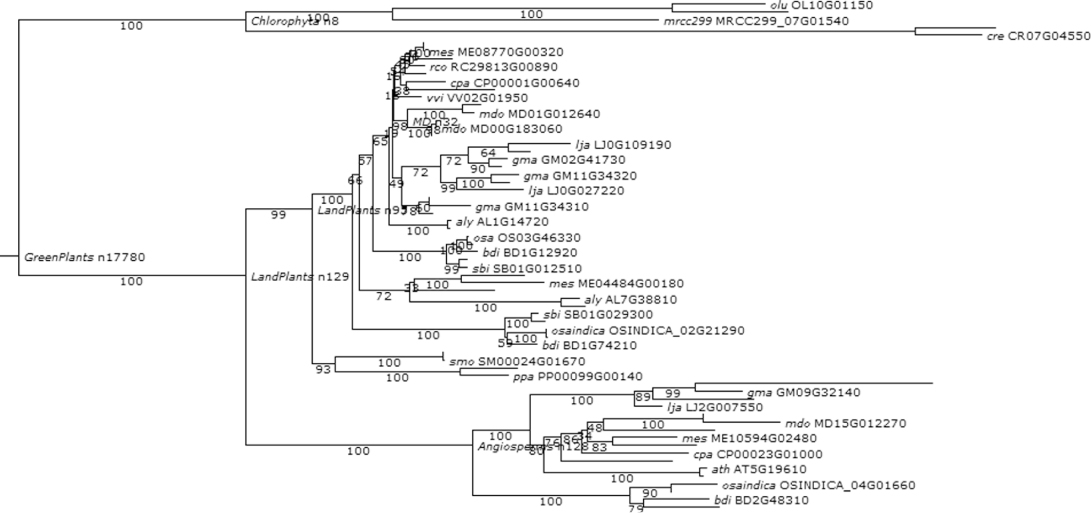
Figure 1.9 Phylogenetic and functional evolutionary history of TORMOZ genes.

Figure 1.10 Phylogenetic and functional evolutionary history of PIN genes.
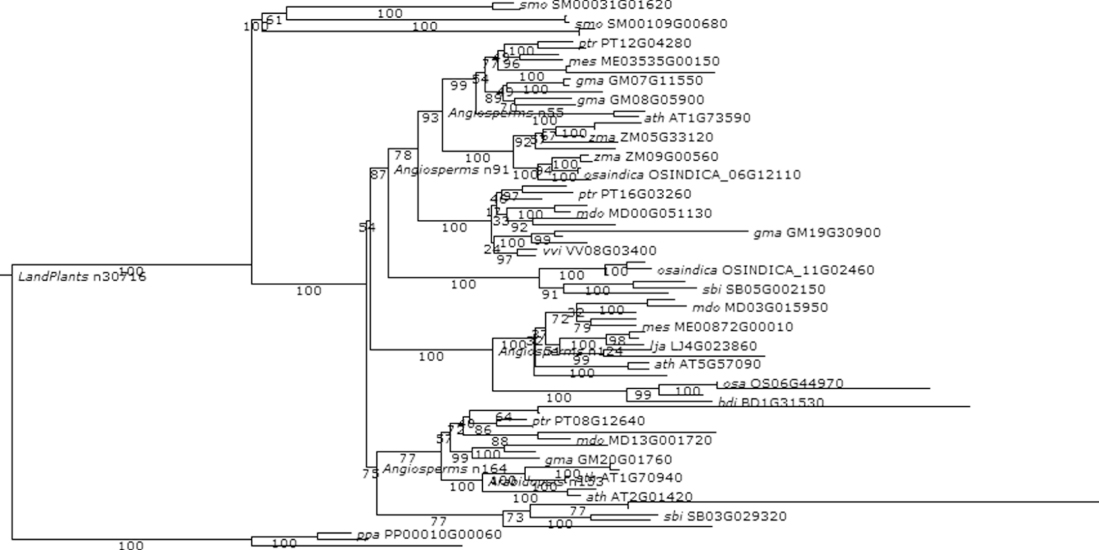
Figure 1.11 Phylogenetic and functional evolutionary history of ACTIN genes.
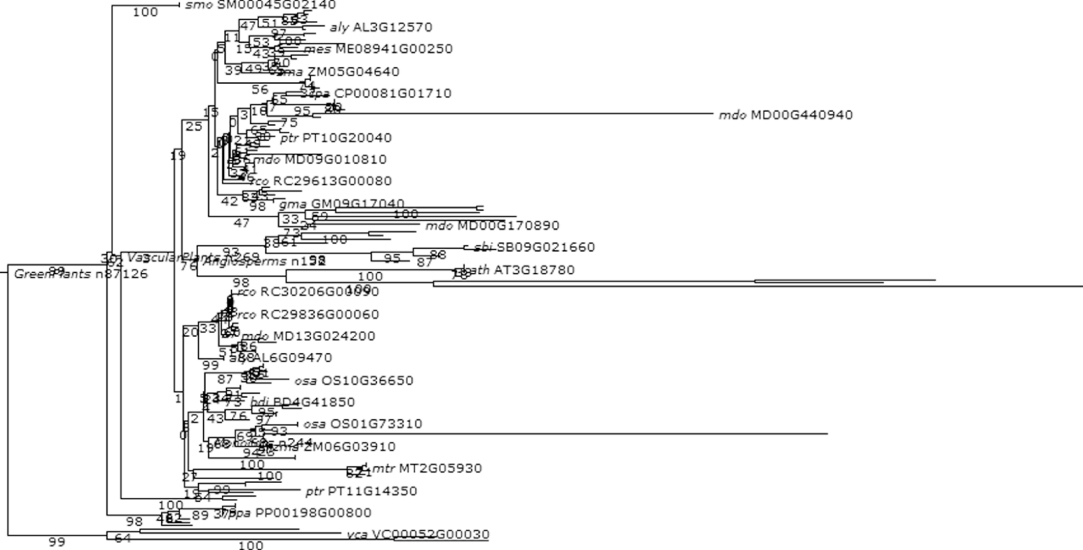
Figure 1.12 Phylogenetic and functional evolutionary history of G proteins.
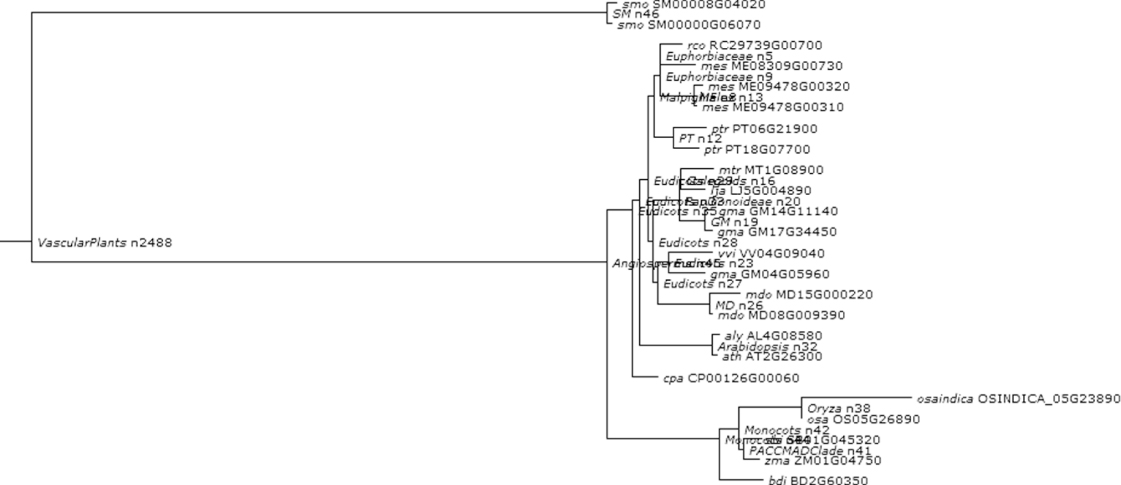
Figure 1.13 Phylogenetic and functional evolutionary history of NPH4TF.
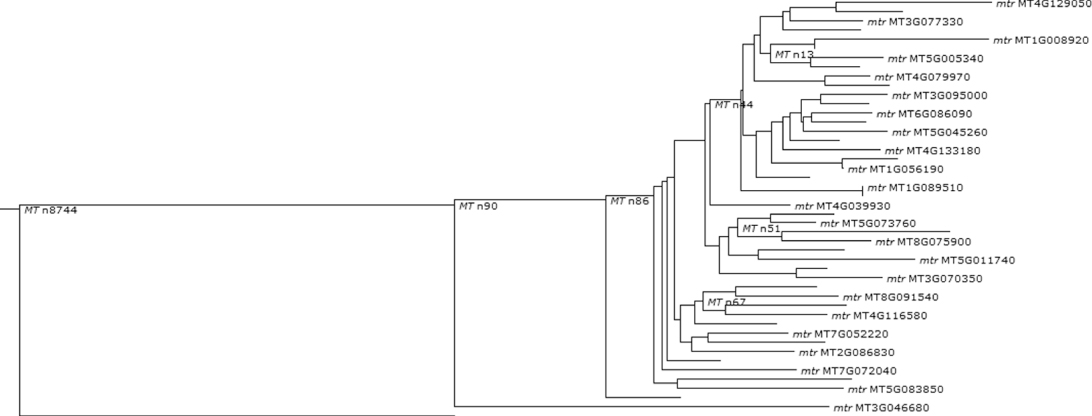
Figure 1.14 Phylogenetic and functional evolutionary history of AGO10.

Figure 1.15 Phylogenetic and functional evolutionary history of LEAFY.
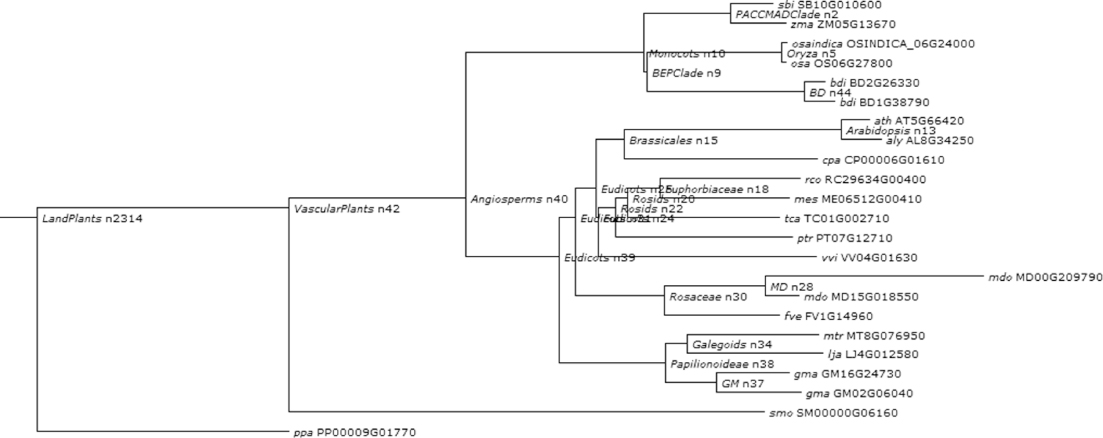
Figure 1.16 Phylogenetic and functional evolutionary history of FT protein.

The following proteins and genes are analyzed here:
1.5 Conclusions
The plant kingdom includes familiar organisms such as trees, herbs, bushes, grasses, vines, ferns, mosses, and green algae. About 350,000 species of plants, defined as seed plants, bryophytes, ferns, and fern allies, are estimated to exist currently. As of 2004, approximately 287,655 species had been identified, of which 258,650 are flowering plants, 16,000 bryophytes, 11,000 ferns, and 8000 green algae. Despite the vast diversity of the plant kingdom in terms of size, shape, and form, nine fundamental body plan features originating from algae have been inherited by all members of the kingdom [5]. The 12 types of biomolecules mentioned in this chapter, are significant for three plant developmental processes such as cell growth, cell differentiation, and morphogenesis. The same molecules have been observed to be related to the nine fundamental body plan features. The relationships between features and types of biomolecules are described as follows:
All these biomolecules, discussed above, play important roles in plant pattern formation. At the same time, these molecules are well conserved along plant evolutionary history (see Table 3).
Table 3 Important Biomelecules and their Biological Roles in Evolutionary History
We further establish the functional and biological significance for selecting these candidate genes in the developmental gene toolkit in plants with PLAZA 2.0 for comparative genomics in plants. This shows the phylogenetic evolution of all those genes consequently. Therefore a genetic toolkit for plant development can be prepared considering all the protein molecules mentioned above as a future project.
References
1. Floyd SK, Bowman JL, The ancestral development toolkit of land plants, Int. J. Plant Sci. 168(1):1–35 (2007).
2. Newman SA, The developmental genetic toolkit and the molecular homology–analogy paradox, Biol. Theory 1(1):12–16 (2006).
3. Kenrick P, Crane PR, The origin and early evolution of plants on land, Nature 389:33–39 (Sept. 2007).
4. Cronk Q, Plant evo-devo (evolution of plant development). Sci. Topics, Aug. 15, 2008 (available at http://www.scitopics.com/Plant_evo_devo_evolution_of_plant_development.html).
5. Graham LE, Cook ME, Busse JS, The origin of plants: Body plan changes contributing to a major evolutionary radiation, Proc. Natl. Acad. Sci. USA 97(9):4535–4540 (April 2000).
6. Srivastava L, Plant Growth and Development: Hormones and Environment, Academic Press, New York, 2002.
7. Whang SS, Confocal microscopy study of Arabidopsis embryogenesis using GFP:mTn, J. Plant Biol. 52(4):312–318 (Aug. 2009).
8. Proseus TE, Boyer SB, Turgor pressure moves polysaccharides into growing cell walls of Chara corallina, Ann. Bot. 95(6):967–979 (2005).
9. Cosgrove DJ, Loosening of plant cell walls by expansins, Nature 407(21):321–326 (Sept. 2000).
10. Fleming AJ, McQueen-Mason S, Mandel T, Kuhlemeier C, Induction of leaf primordia by the cell wall protein expansin, Science 276:1415–1418 (1997).
11. Hall Q, Cannon MC, The cell wall hydroxyproline-rich glycoprotein RSH is essential for normal embryo development in Arabidopsis, Plant Cell 14:1161–1172 (2002).
12. Shevell DE, Leu WM, Gillmor CS, Xia G, Feldman KA, Chua NH, EMB30 is essential for normal cell division, cell expansion, and cell adhesion in Arabidopsis and encodes a protein that has similarity to Sec7, Cell 77:1051–1062 (1994).
13. Bonifacino JS, Jackson CL, Endosome-specific localization and function of the ARF activator GNOM, Cell 112(2):141–142 (2003).
14. Geldner N, Cell polarity in plants: A PARspective on PINs, Curr. Opin. Plant Biol. 12(1):42–48 (2009).
15. Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jürgens G, Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis, Nature 426:147–153 (2003).
16. Gao X, Nagawa S, Wang G, Yang Z, Cell polarity signaling: Focus on polar auxin transport, Mol. Plant 1(6):899–909 (2008).
17. Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B, The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots, Nature 433:39–44 (2005).
18. Feraru E. et al, PIN polar targeting, Plant Physiol. 147:1553–1559 (2008).
19. Dhonukshe P. et al, Generation of cell polarity in plants links endocytosis, auxin distribution and cell fate decisions, Nature 456:962–966 (2008).
20. Christensen SK, Dagenais N, Chory J, Weigel D, Regulation of auxin response by the protein kinase PINOID, Cell 100(4):469–478 (2000).
21. Dhonukshe P, Kleine-Vehn J, Friml J, Cell polarity, auxin transport, and cytoskeleton-mediated division planes: Who comes first? Protoplasma 226(1–2):67–73 (2005).
22. Geldner N, Friml J, Stierhof YD, Jurgens G, Palme K, Auxin transport inhibitors block PIN1 cycling and vesicle trafficking, Nature 413:425–428 (2001).
23. Hou G, Kramer VL, Wang YS, Chen R, Perbal G, Gilroy S, Blancaflor EB, The promotion of gravitropism in Arabidopsis roots upon actin disruption is coupled with the extended alkalinization of the columella cytoplasm and a persistent lateral auxin gradient, Plant J. 39:113–125 (2004).
24. Szymanski DB, Plant cells taking shape: New insights into cytoplasmic control, Curr. Opinion Plant Biol. 12:735–744 (2009).
25. Nick P, Han M-J, An G, Auxin stimulates its own transport by shaping actin filaments, Plant Physiol. 151:155–167 (2009).
26. Mathur J, Martin H, Microtubules and microfilaments in cell morphogenesis in higher plants, Curr. Biol. 12:R669–R676 (2002).
27. Kandasamy MK, McKinney EC, Meagher RB, A single vegetative actin isovariant overexpressed under the control of multiple regulatory sequences is sufficient for normal Arabidopsis development, Plant Cell 21(3):701–718 (2009).
28. Huang S, An YQ, McDowell JM, McKinney EC, Meagher RB, The Arabidopsis ACT11 actin gene is strongly expressed in tissues of the emerging inflorescence, pollen, and developing ovules, Plant Mol. Biol. 33(1):125–139 (1997).
29. Griffith ME, Mayer U, Capron A, Ngo QA, Surendrarao A, McClinton R, Jürgens G, Sundaresan V, The TORMOZ gene encodes a nucleolar protein required for regulated division planes and embryo development in Arabidopsis, Plant Cell 19(7):2246–2263 (2007).
30. Chen J-G, Gao Y, Jones AM, Differential roles of Arabidopsis heterotrimeric G-protein subunits in modulating cell division in roots, Plant Physiol. 141(3):887–897 (2006).
31. Ullah H, Chen J-G, Temple B, Boyes DC, Alonso JM, Davis KR, Ecker JR, Jones AM, The β-subunit of the Arabidopsis G protein negatively regulates auxin-induced cell division and affects multiple developmental processes, Plant Cell 15(2):393–409 (2003).
32. Harper RM, Stowe-Evans EL, Luesse DR, Muto H, Tatematsu K, Watahiki MK, Yamamoto K, Liscum E, The NPH4 locus encodes the auxin response factor ARF7, a conditional regulator of differential growth in aerial Arabidopsis tissue, Plant Cell 12(5):757–770 (2000).
33. Tatematsu K, Kumagai S, Muto H, Sato A, Watahiki MK, Harper RM, Liscum E, Yamamoto KT, MASSUGU2 encodes Aux/IAA19, an auxin-regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypocotyl and formation of lateral roots in Arabidopsis thaliana, Plant Cell 16(2):379–393 (Feb. 2004).
34. Esmon CA, Tinsley AG, Ljung K, Sandberg G, Hearne LB, Liscum E, A gradient of auxin and auxin-dependent transcription precedes tropic growth responses, Proc. Natl. Acad. Sci. 103(1):236–241 (2006).
35. Hutvagner G, Simard MJ, Argonaute proteins: Key players in RNA silencing, Nat. Rev. Mol. Cell. Biol. 9(1):22–32 (2008).
36. Liu Q,Yao X, Pi L, Wang H, Cui X, Huang H, The ARGONAUTE10 gene modulates shoot apical meristem maintenance and establishment of leaf polarity by repressing miR165/166 in Arabidopsis, Plant J. 58(1):27–40 (2009).
37. Teper-Bamnolker P, Samach A, The flowering integrator FT regulates SEPALLATA3 and FRUITFULL accumulation in Arabidopsis leaves, Plant Cell 17(10):2661–2675 (2005).
38. Adrian J, Farrona S, Reimer JJ, Albani MC, Coupland G, Turck F, cis-Regulatory elements and chromatin state coordinately control temporal and spatial expression of flowering locus T in Arabidopsis, Plant Cell 22(5):1425–1440 (May 2010).
39. Notaguchi M, Abe M, Kimura T, Daimon Y, Kobayashi T, Yamaguchi A, Tomita Y, Dohi K, Mori M, Araki T, Long-distance, graft-transmissible action of Arabidopsis flowering locus T protein to promote flowering, Plant Cell Physiol. 49:1645–1658 (2008).
40. Wu X, Weigel D, Wigge PA, Signaling in plants by intercellular RNA and protein movement, Genes Dev. 16:151–158 (2002).
41. Haywood V, Kragler F, Lucas WJ, Plasmodesmata pathways for protein and ribonucleoprotein signaling, Plant Cell 14:S303–S325 (May 2002).
42. Winter CM, Austin RS, Reback MA, Wu M, Yamaguchi A, Li H, Wagner D, LEAFY target genes reveal a direct link between external stimulus response and flower development, Proc. 21st Int. Conf. Arabidopsis Research, Japan 2010.
43. Moyroud E, Kusters E, Monniaux M, Koes R, Parcy F, LEAFY blossoms, Trends Plant Sci. 15(6):346–352 (June 2010).
44. Maizel A, Busch MA, Tanahashi T, Perkovic J, Kato M, Hasebe M, Weigel D, The floral regulator LEAFY evolves by substitutions in the DNA binding domain, Science 308(5719):260 (2005).
45. Liu C, Thong Z, Yu H, Coming into bloom: the specification of floral meristems. Development 136:3379–3391 (2009). doi:10.1242/dev.033076.
46. Kato T, Morita MT, Tasaka M, Defects in dynamics and functions of actin filament in Arabidopsis caused by the dominant-negative actin fiz1-induced fragmentation of actin filament, Plant Cell Physiol. 51(2):333–338 (2010). doi:10.1093/pcp/pcp189.
47. Sessions A, Yanofsky MF, Weigel D, Cell-cell signaling and movement by the floral transcription factors LEAFY and APETALA1, Science 289(5480):779 (2000). doi: 10.1126/science.289.5480.779.
48. Kenrick P, Crane P, The Origin and Early Diversification of Land Plants: A Cladistic Study. Smithsonian Institution Press, Washington, DC, 1997.
49. Hamilton A, Hamilton P, Plant Conservation: An Ecosystem Approach, Earthscan, London, 2006, p. 2.
50. Wang H-C, Moore MJ, Soltis PS, Bell CD, Brockington SF, Alexandre R, Davis CC, Latvis M, Manchester SR, Soltis DE, Rosid radiation and the rapid rise of angiosperm-dominated forests, Proceedings of the National Academy of Sciences 106(10):3853–3858 (2009).
51. Scotland RW, Wortley AH, How many species of seed plants are there?, Taxon 52(1):101–104 (2003).
52. Soltis DE, Soltis PS, Endress PK, Chase MW, Phylogeny and Evolution of the Angiosperms, Sinauer, Sunderland, MA, 2005.
53. Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, Rokhsar DS, Phytozome: a comparative platform for green plant genomics, Nucleic Acids Res. 40(D1):D1178–D1186 (2012).
54. Proost S, Van Bel M, Sterck L, Billiau K, Van Parys T, Van de Peer Y, Vandepoele K, PLAZA: a comparative genomics resource to study gene and genome evolution in plants, Plant Cell 21:3718–3731 (2009).
Bibliography
Cannon MC, Terneus K, Hall Q, Tan L, Wang Y, Wegenhart BL, Chen L, Lamport DT, Chen Y, Kieliszewski MJ, Self-assembly of the plant cell wall requires an extensin scaffold, Proc. Natl. Acad. Sci. USA 105(6):2226–2231 (2008).
Corbesier L, Vincent C, Jang SH, Fornara F, Fan QZ, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, Coupland G, FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis, Science 316:1030–1033 (2007).
Donaldson JG, Jackson CL, A cell-centered approach to developmental biology, Curr. Opin. Cell Biol. 12:475–482 (2000).
Dupree P, Plant embryogenesis: Cell division forms a pattern, Curr. Biol. 6(6):683–685 (June 1996).
Geldner N, The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth, Cell 112:219–230 (2003).
Jaeger KE, Wigge PA, FT protein acts as a long-range signal in Arabidopsis, Curr. Biol. 17:1050–1054 (2007).
Jürgens G, Apical-basal pattern formation in the Arabidopsis embryo: Studies on the role of the gnom gene, Development 117:149–162 (1993).
Jürgens G, Apical-basal pattern formation in Arabidopsis embryogenesis, EMBO J. 20:3609–3616 (2001).
Kleine-Vehn J, Dhonukshe P, Swarup R, Bennett M, Friml J, Subcellular trafficking of the Arabidopsis auxin influx carrier AUX1 uses a novel pathway distinct from PIN1, Plant Cell 18(11):3171–3181 (2006).
Kleine-Vehn J, Langowski L, Wisniewska J, Dhonukshe P, Brewer PB, Friml J, Cellular and molecular requirements for polar PIN targeting and transcytosis in plants, Mol. Plant 1(6):1056–1066 (2008).
Kleine-Vehn J, Friml J, Polartargeting and endocytic recycling in auxin-dependent plant development, Annu. Rev. Cell Dev. Biol. 24:447–473 (2008).
Kleine-Vehn J, Dhonukshe P, Sauer M, Brewer PB, Wisniewska J, Paciorek T, Benkova E, Friml J, ARF GEF-dependent transcytosis and polar delivery of PIN auxin carriers in Arabidopsis, Curr. Biol. 18:526–531 (2008).
Lyndon RF, Plant Development: The Cellular Basis, Unwin Hyman, London, 1990.
Mayer U, Büttner G, Jürgens G, Apical-basal pattern formation in the Arabidopsis embryo: Studies on the role of the gnom gene, Development 117:149–162 (1993).
Ma H, Plant G proteins: The different faces of GPA1, Curr. Biol. 11:R869–R871 (2001).
Merks R, Glazier J, Statistical mechanics and its applications, Phys A. Appl. Biol. Syst. 352(1):113–130 (July 2005).
Mathieu J, Warthmann N, Kuttner F, Schmid M, Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis, Curr. Biol. 17:1055–1060 (2007).
Niklas KJ, Simulation of organic shape: The roles of phenomenology and mechanism, J. Morphol. 219(3):243–246 (2005).
Niklas KJ, Spatz H-C, Vincent J, Plant biomechanics: An overview and prospectus, Am. J. Bot. 93:1369–1378 (2006).
Perrot-Rechenmann C, Cellular responses to auxin: Division versus expansion, Cold Spring Harbor Perspect. Biol. 2(5):a001446 (May 2010).
Salazar-Ciudad I, Looking at the origin of phenotypic variation from pattern formation gene networks, J. Biosci. 34:573–587 (2009).
Schopfer P, Biomechanics of plant growth, Am. J. Bot. 93:1415–1425 (2006).
Steinmann T, Geldner N, Grebe M, Mangold S, Jackson CL, Paris S, Galweiler L, Palme K, Jürgens G , Coordinated polar localization of auxin efflux carrier PIN1 by GNOM ARF GEF, Science 286(5438):316–318 (1999).
Surpin M, Raikhel N, Traffic jams affect plant development and signal transduction, Nature 5:100–109 (2004).
Zhang J, Nodzynski T, Pencik A, Rolcik J, Friml J, PIN phosphorylation is sufficient to mediate PIN polarity and direct auxin transport, Proc. Natl. Acad. Sci. 107:918–922 (2010).
