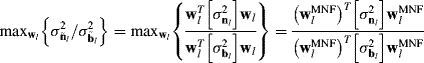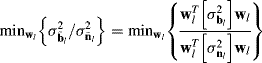6.2 Dimensionality Reduction by Second-Order Statistics-Based Component Analysis Transforms
A CA transform generally transforms the image data into a set of data components so that the correlation among the transformed data components is uncorrelated according to a criterion. More specifically, a component transform represents a data space by a set of its generated data components. Two second-order component transforms have been widely used in remote sensing image processing, which are variance-based PCA transforms and SNR-based transforms and discussed as follows.
6.2.1 Eigen Component Analysis Transforms
The simplest eigen-CA transforms are those based on data variance. PCA represents this type of data variance-based CA transforms.
6.2.1.1 Principal Components Analysis
The principal components analysis (PCA), also known as Hotelling transform (Gonalez and Woods, 2002) as well as principal components transformation (PCT) (Richards and Jia, 1999; Schowengerdt, 1997), is an optimal transform to represent data in the sense of data variance. It can be considered as a discrete time version of the Karhunen–Loeve transform (KLT) in signal processing and communications (Poor, 1994) that is an optimal transform using eigenfunctions as basis functions to represent and de-correlate a function in the sense of mean-squared error. It is generally referred to as Karhunen–Loeve expansion that represents a function as a series in terms of eigenfunctions where these eigenfunctions are continuous-time functions. When they are sampled at discrete time instants, eigenfunctions become eigenvectors in a discrete case, in which case KLT is reduced to PCA. So, technically speaking, KLT used in hyperspectral data compression is indeed the principal components analysis (PCA) not what it was originally developed in statistical signal processing and communications in at least two major key aspects. The first and foremost is the used criterion. While KLT is a mean-squared error (MSE)-based transform that assumes the availability of data probability distribution to perform “mean” in terms of statistical expectation, PCA makes use of the sample covariance matrix without assuming the data probability distribution in which case PCA should be considered as a “least squares error (LSE)”-based transform, not an MSE-based transform where LSE is actually the sample variance. Secondly, KLT is generally referred to as KL expansion in statistical signal processing where a signal can be decomposed as a series of orthogonal functions, to be called eigenfunctions. For example, Fourier transform is a special case of KLT where the used sinusoidal functions are basically eigenfunctions. Therefore, in general, KLT is a continuous time transform function. By contrast, PCA is a matrix transform used to de-correlate data sample vectors into linear combinations of using eigenvectors as basis vectors for their data representations with eigenvalues as their corresponding coefficients. In light of this interpretation PCA is indeed a discrete time of KL expansion. Unfortunately, KLT has been widely abused in image processing where the image data are represented by matrices in which case KLT should be implemented as its discrete-time version, PCA. It seems that such key differences have been overlooked in hyperspectral data compression. The idea of PCA can be briefly described as follows.
Assume that ![]() is a set of L-dimensional image pixel vectors and μ is the mean of the sample pool S obtained by
is a set of L-dimensional image pixel vectors and μ is the mean of the sample pool S obtained by ![]() . Let X be the sample data matrix formed by
. Let X be the sample data matrix formed by ![]() . Then the sample covariance matrix of the S is obtained by
. Then the sample covariance matrix of the S is obtained by ![]() . If we further assume that
. If we further assume that ![]() is the set of eigenvalues obtained from the covariance matrix K and
is the set of eigenvalues obtained from the covariance matrix K and ![]() are their corresponding unit eigenvectors, that is,
are their corresponding unit eigenvectors, that is, ![]() , we can define a diagonal matrix Dσ with variances
, we can define a diagonal matrix Dσ with variances ![]() along the diagonal line as
along the diagonal line as
(6.1) 
and an eigenvector matrix Λ specified by ![]() as
as
such that
Using the eigenvector matrix Λ a linear transform ξΛ defined by
transforms every data sample ri to a new data sample, ![]() by
by
As a result, the mean of new ξΛ-transferred data samples ![]() becomes
becomes ![]() and its resulting covariance matrix is reduced to a diagonal matrix given by
and its resulting covariance matrix is reduced to a diagonal matrix given by
Equation (6.6) implies that the ξΛ-transferred data matrix ![]() has been de-correlated or whitened by the matrix Λ that is referred to as a whitening matrix (Poor, 1994). The transform ξΛ defined by (6.5) is generally called principal component transform and the lth component of
has been de-correlated or whitened by the matrix Λ that is referred to as a whitening matrix (Poor, 1994). The transform ξΛ defined by (6.5) is generally called principal component transform and the lth component of ![]() is formed by
is formed by
and is called the lth principal component (PC) that consists of ![]() that are
that are ![]() -transferred data samples corresponding the lth eigenvalue λl. PCA is a process that implements the transform ξΛ defined by (6.4) to obtain a set of principal components (PCs) via (6.5) or (6.7) with all
-transferred data samples corresponding the lth eigenvalue λl. PCA is a process that implements the transform ξΛ defined by (6.4) to obtain a set of principal components (PCs) via (6.5) or (6.7) with all ![]() . In order to achieve DR, only the PCs specified by eigenvectors that correspond to first q largest eigenvalues will be retained, while the PCs specified by eigenvectors corresponding to the remaining (L–q) smaller eigenvalues will be discarded. The same process can be accomplished by the singular value decomposition (SVD) to be described in Section 6.2.1.6.
. In order to achieve DR, only the PCs specified by eigenvectors that correspond to first q largest eigenvalues will be retained, while the PCs specified by eigenvectors corresponding to the remaining (L–q) smaller eigenvalues will be discarded. The same process can be accomplished by the singular value decomposition (SVD) to be described in Section 6.2.1.6.
6.2.1.2 Standardized Principal Components Analysis
In PCA, its focus is placed on the variances of the image pixel vectors ![]() . It has been shown by Singh and Harison (1985) that in some applications in remote sensing, it may be more effective to deal with data co-variances rather than data variances. Such co-variance-based PCA is called standardized principal components analysis (SPCA).
. It has been shown by Singh and Harison (1985) that in some applications in remote sensing, it may be more effective to deal with data co-variances rather than data variances. Such co-variance-based PCA is called standardized principal components analysis (SPCA).
Assume that the covariance matrix K is given by
(6.8) 
with the lth variance and (i,j)-covariance denoted by ![]() and σij, respectively. Now we define a standard deviation matrix of K via (6.78) as
and σij, respectively. Now we define a standard deviation matrix of K via (6.78) as ![]() that is the diagonal matrix of the form
that is the diagonal matrix of the form
Then ![]() is called a standardized sample of ri and K can be expressed as
is called a standardized sample of ri and K can be expressed as
where RK is called the correlation coefficient matrix defined by
with ![]() and
and ![]() . It should be noted that the RK in (6.11) is not the sample correlation matrix R. The κij in RK is generally called the (i,j)th correlation coefficient of K.
. It should be noted that the RK in (6.11) is not the sample correlation matrix R. The κij in RK is generally called the (i,j)th correlation coefficient of K.
Using (6.2)![]() is the eigenvector matrix of K formed by its unit eigenvectors
is the eigenvector matrix of K formed by its unit eigenvectors ![]() . Through (6.10) we can obtain
. Through (6.10) we can obtain
that is the ![]() identity matrix. Combining the eigenvector matrix Λ in (6.2) and the diagonal matrix
identity matrix. Combining the eigenvector matrix Λ in (6.2) and the diagonal matrix ![]() obtained by (6.9) we can define a linear transform
obtained by (6.9) we can define a linear transform ![]() by
by
(6.13) ![]()
that is called standardized PCA (SPCA) and denoted by
Using (6.12) and (6.14), the covariance matrix of the new data samples ![]() that are obtained from
that are obtained from ![]() by the SPCA in (6.14) becomes an identity matrix.
by the SPCA in (6.14) becomes an identity matrix.
Similarly, in analogy with the decomposition of K, its inverse matrix K−1 can be also decomposed as
(6.15) ![]()
where
and ![]() are variances of K−1 and
are variances of K−1 and
with ηij being the (i,j)th correlation coefficient of K−1 and ![]() . It turns out that the
. It turns out that the ![]() in (6.16) can be related to the
in (6.16) can be related to the ![]() in (6.9) by the following formula:
in (6.9) by the following formula:
where ![]() is a multiple correlation coefficient of the data in the lth dimension on all other
is a multiple correlation coefficient of the data in the lth dimension on all other ![]() dimensions obtained by using the multiple regression theory. So,
dimensions obtained by using the multiple regression theory. So, ![]() is the reciprocal of a good noise variance estimate for the lth-dimensional data space. It should be noted that the Dζ in (6.16) is not an inverse of the Dσ in (6.9), nor is
is the reciprocal of a good noise variance estimate for the lth-dimensional data space. It should be noted that the Dζ in (6.16) is not an inverse of the Dσ in (6.9), nor is ![]() in (6.17). The major advantage of using ζl over
in (6.17). The major advantage of using ζl over ![]() is that as shown in (6.18), ζl removes its correlation on other ζl's for
is that as shown in (6.18), ζl removes its correlation on other ζl's for ![]() , while
, while ![]() does not. Like PCA, SPCA achieves DR by only retaining standard PCs corresponding to eigenvectors that are associated with first q largest eigenvalues.
does not. Like PCA, SPCA achieves DR by only retaining standard PCs corresponding to eigenvectors that are associated with first q largest eigenvalues.
6.2.1.3 Singular Value Decomposition
Another eigen-CA transform is the singular value decomposition (SVD). Unlike PCA that is primarily designed to de-correlate the covariance matrix, SVD is one of most widely used techniques in systems, communications, and signal processing to resolve issues caused by ill-conditioned systems, such as underdetermined or overdetermined least squares system. It provides a matrix factorization of an arbitrary matrix into a product of two unitary matrices and a diagonal matrix. More specifically, let ![]() be an
be an ![]() real matrix. Define two matrices
real matrix. Define two matrices ![]() and
and ![]() that can, respectively, be referred to as outer product matrix and inner product matrix where both matrices
that can, respectively, be referred to as outer product matrix and inner product matrix where both matrices ![]() and
and ![]() are symmetric, semidefinite with non-negative real eigenvalues, and have the same rank. In particular, when
are symmetric, semidefinite with non-negative real eigenvalues, and have the same rank. In particular, when ![]() is an m-dimensional column vector x, the outer product matrix is an
is an m-dimensional column vector x, the outer product matrix is an ![]() matrix,
matrix, ![]() , and the inner product matrix
, and the inner product matrix ![]() is a scalar, both of which have rank 1. In this case, the only nonzero eigenvalue of the outer product matrix of
is a scalar, both of which have rank 1. In this case, the only nonzero eigenvalue of the outer product matrix of ![]() is specified by its inner product matrix
is specified by its inner product matrix ![]() .
.
Assume that the eigenvalues of ![]() and
and ![]() are
are ![]() and
and ![]() that can be arranged in descending order in magnitude as follows:
that can be arranged in descending order in magnitude as follows:
where ![]() . Since both matrices
. Since both matrices ![]() and
and ![]() have the same rank and also identical nonzero eigenvalues,
have the same rank and also identical nonzero eigenvalues, ![]() and
and ![]() for all
for all ![]() . Then the set of square root of eigenvalues
. Then the set of square root of eigenvalues ![]() in (6.19)
in (6.19)
(6.20) ![]()
is called the singular values of the matrix ![]() (Chen, 1999). Now we can further decompose the matrix
(Chen, 1999). Now we can further decompose the matrix ![]() into the following factorization form:
into the following factorization form:
where ![]() is an
is an ![]() unitary matrix with its column vectors being orthonormalized eigenvectors of the
unitary matrix with its column vectors being orthonormalized eigenvectors of the ![]() matrix
matrix ![]() so that
so that ![]() ,
, ![]() is an
is an ![]() unitary matrix with its column vectors being orthonormalized eigenvectors of the
unitary matrix with its column vectors being orthonormalized eigenvectors of the ![]() matrix
matrix ![]() so that
so that ![]() , and
, and ![]() is an
is an ![]() diagonal matrix
diagonal matrix ![]() with its diagonal entries specified by the singular values of
with its diagonal entries specified by the singular values of ![]() and arranged in descending order in magnitude,
and arranged in descending order in magnitude, ![]() . Specifically, if the rank of
. Specifically, if the rank of ![]() is r, then
is r, then ![]() is a square matrix of size
is a square matrix of size ![]() with
with ![]() .
.
In hyperspectral data exploitation, assume that ![]() is a set of entire image pixel vectors or a set of training samples in a hyperspectral image.
is a set of entire image pixel vectors or a set of training samples in a hyperspectral image. ![]() in (6.21) can be considered by either a data matrix formed by data samples/image pixel vectors with the subscript m and n denoting the total number of spectral bands and the number of image pixel vectors (such as total number of image pixel vectors or training samples), respectively, or a sample correlation/covariance matrix
in (6.21) can be considered by either a data matrix formed by data samples/image pixel vectors with the subscript m and n denoting the total number of spectral bands and the number of image pixel vectors (such as total number of image pixel vectors or training samples), respectively, or a sample correlation/covariance matrix ![]() /
/![]() formed by the total number of data samples/image pixel vectors with
formed by the total number of data samples/image pixel vectors with ![]() being the total number of spectral bands, and q being the number of dimensions to be retained, respectively. In the former case, the matrix
being the total number of spectral bands, and q being the number of dimensions to be retained, respectively. In the former case, the matrix ![]() in (6.21) is formed by data sample vectors,
in (6.21) is formed by data sample vectors, ![]() with the ith column vector specified by the ith image pixel vector ri. So, the resulting matrix is represented by
with the ith column vector specified by the ith image pixel vector ri. So, the resulting matrix is represented by ![]() with
with ![]() . In (6.21)
. In (6.21) ![]() and
and ![]() are called left and right singular vector matrices of
are called left and right singular vector matrices of ![]() and they are generally different. The singular values of
and they are generally different. The singular values of ![]() are simply square root of non-negative eigenvalues of
are simply square root of non-negative eigenvalues of ![]() ,
, ![]() . In other words, if we interpret eigenvalues as variances, the singular values are simply their standard deviations. As for the latter case, the matrix
. In other words, if we interpret eigenvalues as variances, the singular values are simply their standard deviations. As for the latter case, the matrix ![]() in (6.21) is formed by the data sample correlation matrix,
in (6.21) is formed by the data sample correlation matrix, ![]() , that becomes the outer product matrix of
, that becomes the outer product matrix of ![]() ,
, ![]() scaled by a constant
scaled by a constant ![]() . The singular values of
. The singular values of ![]() are exactly the same non-negative eigenvalues of
are exactly the same non-negative eigenvalues of ![]() , and the left and right singular vector matrices of
, and the left and right singular vector matrices of ![]() ,
, ![]() , and
, and ![]() turn out to be the same as the eigenvector matrix Λ described by (6.2); (6.21) is reduced to (6.3), in which case SVD becomes PCA described in Section 6.2.1.1.
turn out to be the same as the eigenvector matrix Λ described by (6.2); (6.21) is reduced to (6.3), in which case SVD becomes PCA described in Section 6.2.1.1.
In order to further explore insights into the relationship between the SVD and PCA, let ![]() and
and ![]() be eigenvalues of the sample covariance matrix
be eigenvalues of the sample covariance matrix ![]() and the sample correlation matrix
and the sample correlation matrix ![]() with their corresponding eigenvectors
with their corresponding eigenvectors ![]() and
and ![]() , respectively. Also let
, respectively. Also let ![]() ,
, ![]() and
and ![]() be the singular values of
be the singular values of ![]() ,
, ![]() and
and ![]() with their corresponding singular vectors
with their corresponding singular vectors ![]() ,
, ![]() and
and ![]() , respectively. The following relationships can be derived and summarized as follows.
, respectively. The following relationships can be derived and summarized as follows.
Finally, as an alternative, we can also find the singular values of the inner product matrix of the matrix ![]() ,
, ![]() with size of
with size of ![]() . It turns out that both inner product matrix of
. It turns out that both inner product matrix of ![]() ,
, ![]() and the outer product matrix of
and the outer product matrix of ![]() ,
, ![]() have the same identical non zero singular values with only difference in the number of zero singular values. This implies that to perform DR for any matrix
have the same identical non zero singular values with only difference in the number of zero singular values. This implies that to perform DR for any matrix ![]() , either inner product matrix
, either inner product matrix ![]() or outer product matrix
or outer product matrix ![]() can be used for SVD. Apparently, in hyperspectral imaging the data sample correlation matrix
can be used for SVD. Apparently, in hyperspectral imaging the data sample correlation matrix ![]() that is a
that is a ![]() -scaled outer product matrix of a data matrix
-scaled outer product matrix of a data matrix ![]() is the most intuitive and logical way to be chosen for DR.
is the most intuitive and logical way to be chosen for DR.
There are also other factorization forms similar to (6.6) that can be used in place of SVD, for example, Cholesky decomposition, QR decomposition, and Householder transformation (Golub and Van Loan, 1989), that can be used for real-time implementation (see Chapter 33 and Chang (2013)).
6.2.2 Signal-to-Noise Ratio-Based Components Analysis Transforms
The PCA discussed in Section 6.2.1 is developed to arrange PCs in descending order of data variance. However, data variance does not mean image quality. In other words, PCA-ordered PCs are not necessarily ordered by image quality as shown by Green et al. (1988). In order to address this issue, Green et al. (1998) used an approach similar to PCA, called maximum noise fraction (MNF), that was based on a different criterion, signal-to-noise (SNR), to measure image quality. It was later shown by Lee et al. (1990) that MNF actually performed two stage processes, noise whitening with unit variance followed by PCA. Because of that MNF was also referred to as noise-adjusted principal component (NAPC) transform.
6.2.2.1 Maximum Noise Fraction Transform
The idea of MNF can be briefly described as follows. Assume that ![]() is a set of entire image pixel vectors in a hyperspectral image with size
is a set of entire image pixel vectors in a hyperspectral image with size ![]() when nr and nc denote the number of rows and columns in the image, respectively. Let each image pixel vector also be denoted by an L-dimensional column vector
when nr and nc denote the number of rows and columns in the image, respectively. Let each image pixel vector also be denoted by an L-dimensional column vector ![]() . Suppose that the lth band image can be represented by an N-dimensional column vector,
. Suppose that the lth band image can be represented by an N-dimensional column vector, ![]() . It assumes that an observation model
. It assumes that an observation model
(6.22) ![]()
where bl is an observation vector, sl is an N-dimensional signal column vector, and nl is an N-dimensional column vector uncorrelated with sl.
Let ![]() and
and ![]() denote the noise variance and signal variance of bl, respectively. We define the noise fraction (NF) of the lth band image vector bl to be the ratio of the noise variance,
denote the noise variance and signal variance of bl, respectively. We define the noise fraction (NF) of the lth band image vector bl to be the ratio of the noise variance, ![]() in the lth band image to the total variance,
in the lth band image to the total variance, ![]() in the lth band image given by
in the lth band image given by
where ![]() and
and ![]() .
.
Assume that wl is an L-dimensional column vector that will be used to linearly transform the lth band image vector ![]() to a new lth band image described by
to a new lth band image described by ![]() via
via
It is worth noting that the ith component of the lth band image vector ![]() ,
, ![]() in (6.24) is obtained by weighted sum over image pixels in all the L bands of the ith image pixel vector ri. So, MNF is to find a transform specified by
in (6.24) is obtained by weighted sum over image pixels in all the L bands of the ith image pixel vector ri. So, MNF is to find a transform specified by ![]() to maximize the NF defined by
to maximize the NF defined by
Let ![]() be an MNF transform matrix such that
be an MNF transform matrix such that ![]() where
where ![]() and
and ![]() . Then we can obtain the lth MNF-transformed band image by
. Then we can obtain the lth MNF-transformed band image by ![]() via
via ![]() specified by (6.24). Since the criterion of NF given by (6.23) can be re-expressed as
specified by (6.24). Since the criterion of NF given by (6.23) can be re-expressed as
where ![]() is signal-to-noise ratio defined in (6.25). As a consequence, maximizing the NFl specified by (6.23) is equivalent to minimizing SNRl specified by (6.26). The Green et al. developed MNF is to find a set of
is signal-to-noise ratio defined in (6.25). As a consequence, maximizing the NFl specified by (6.23) is equivalent to minimizing SNRl specified by (6.26). The Green et al. developed MNF is to find a set of ![]() to maximize the noise fraction in each of bands and then arrange MNF-transformed bands in descending order of maximum noise fractions according to (6.23) or in ascending order of SNR according to (6.26).
to maximize the noise fraction in each of bands and then arrange MNF-transformed bands in descending order of maximum noise fractions according to (6.23) or in ascending order of SNR according to (6.26).
6.2.2.2 Noise-Adjusted Principal Component Transform
Recently, Lee et al. (1990) re-interpreted MNF transform and showed that MNF transform was nothing more than a two-stage process that first whitened noise variances of each band image to unit variance, then performed PCA transform on the noise-whitened band images. As a result, PCA-generated principal components can be arranged in the descending order of SNR that is the reverse order arranged by MNF transform. With this new interpretation, MNF is further referred to as noise-adjusted principal component (NAPC) transform. In other words, we can reinterpret MNF transform that maximizes the NFl in (6.23) to minimize its reciprocal defined by
or maximize the SNR over the reciprocal of (6.26) defined by
As a result of (6.27) or (6.28), the obtained transform vectors arrange band images in ascending order of noise fractions or descending order of SNR. Interestingly, MNF used in the popular ENVI software is actually minimum noise fraction specified by (6.27).
The argument outlined above by (6.27) and (6.28) was based on Green et al.'s approach for each band image bl, not an entire hyperspectral image cube. As noted, the lth MNF-transformed band image vector ![]() is obtained by (6.24) whose ith component
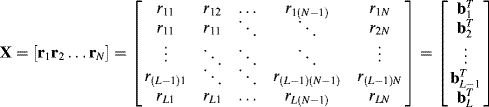
is obtained by (6.24) whose ith component ![]() is actually calculated by a weighted band correlation among the L bands within the ith image pixel vector via a particular weight vector wl. It may not be conceptually clear and easy to be understood from a hyperspectral image viewpoint as an image cube. However, the connection between Green et al.'s MNF transform and Lee et al.'s NAPC can be better understood if a hyperspectral image is presented as a data matrix as follows. Following the same notations used in the MNF transform, assume that an L-band hyperspectral image has N image pixels denoted by
is actually calculated by a weighted band correlation among the L bands within the ith image pixel vector via a particular weight vector wl. It may not be conceptually clear and easy to be understood from a hyperspectral image viewpoint as an image cube. However, the connection between Green et al.'s MNF transform and Lee et al.'s NAPC can be better understood if a hyperspectral image is presented as a data matrix as follows. Following the same notations used in the MNF transform, assume that an L-band hyperspectral image has N image pixels denoted by![]() with
with ![]() where nr and nc denote the number of rows and columns in the image, respectively. Also, let
where nr and nc denote the number of rows and columns in the image, respectively. Also, let ![]() be an N-dimensional column vector that represents the lth band image of the hyperspectral image. Then the relationship between L-dimensional image pixel vectors
be an N-dimensional column vector that represents the lth band image of the hyperspectral image. Then the relationship between L-dimensional image pixel vectors ![]() ri and L band images
ri and L band images ![]() can be related by the following data matrix
can be related by the following data matrix ![]() :
:
and
According to (6.29) and (6.30), Green et al.'s MNF transform performs on the left-hand side of (6.30) band-by-band images in a similar fashion as a remotely sensed image is stored by the Band SeQuential (BSQ) (Schowengerdt, 1997, p. 25). On the other hand, Lee et al.'s NAPC transform processes a hyperspectral image as the data matrix, that is, ![]() on the left-hand side of (6.29) in the same way as a remotely sensed image is stored by the band-interleaved-by-pixel (BIP) (Schowengerdt, 1997, p. 26). Therefore, in the NAPC transform, the sample data covariance matrix is obtained by
on the left-hand side of (6.29) in the same way as a remotely sensed image is stored by the band-interleaved-by-pixel (BIP) (Schowengerdt, 1997, p. 26). Therefore, in the NAPC transform, the sample data covariance matrix is obtained by ![]() and the noise covariance matrix, Kn is estimated from the data matrix X (Lee et al. 1990). A fast algorithm derived by Roger (1994) to implement NAPC transform is summarized as follows.
and the noise covariance matrix, Kn is estimated from the data matrix X (Lee et al. 1990). A fast algorithm derived by Roger (1994) to implement NAPC transform is summarized as follows.
Algorithm for NAPC Transform
(6.31) ![]()
(6.32) ![]()
According to (6.25) and (6.33), MNF transform and NAPC transform achieve DR by only retaining first q projection vectors ![]() and
and ![]() that correspond to the q largest SNRs
that correspond to the q largest SNRs
One major disadvantage of implementing MNF or NAPC transform is estimation of noise covariance matrix. Since it is based on the criterion of SNR, reliable noise estimation must be guaranteed. For details of estimation of noise covariance matrix we refer to Section 17.3 in Chang (2003a).