Apache Spark is an evolution of Hadoop and has become very popular in the last few years. Contrarily to Hadoop and its Java and batch-focused design, Spark is able to produce iterative algorithms in a fast and easy way. Furthermore, it has a very rich suite of APIs for multiple programming languages and natively supports many different types of data processing (machine learning, streaming, graph analysis, SQL, and so on).
Apache Spark is a cluster framework designed for quick and general-purpose processing of big data. One of the improvements in speed is given by the fact that data, after every job, is kept in-memory and not stored on the filesystem (unless you want to) as would have happened with Hadoop, MapReduce, and HDFS. This thing makes iterative jobs (such as the clustering K-means algorithm) faster and faster as the latency and bandwidth provided by the memory are more performing than the physical disk. Clusters running Spark, therefore, need a high amount of RAM memory for each node.
Although Spark has been developed in Scala (which runs on the JVM, like Java), it has APIs for multiple programming languages, including Java, Scala, Python, and R. In this book, we will focus on Python.
Spark can operate in two different ways:
- Standalone mode: It runs on your local machine. In this case, the maximum parallelization is the number of cores of the local machine and the amount of memory available is exactly the same as the local one.
- Cluster mode: It runs on a cluster of multiple nodes, using a cluster manager such as YARN. In this case, the maximum parallelization is the number of cores across all the nodes composing the cluster and the amount of memory is the sum of the amount of memory of each node.
In order to use the Spark functionalities (or pySpark, containing the Python APIs of Spark), we need to instantiate a special object named SparkContext. It tells Spark how to access the cluster and contains some application-specific parameters. In the IPython Notebook provided in the virtual machine, this variable is already available and named sc (it's the default option when an IPython Notebook is started); let's now see what it contains.
First, open a new IPython Notebook; when it's ready to be used, type the following in the first cell:
In:sc._conf.getAll() Out:[(u'spark.rdd.compress', u'True'), (u'spark.master', u'yarn-client'), (u'spark.serializer.objectStreamReset', u'100'), (u'spark.yarn.isPython', u'true'), (u'spark.submit.deployMode', u'client'), (u'spark.executor.cores', u'2'), (u'spark.app.name', u'PySparkShell')]
It contains multiple information: the most important is the spark.master, in this case set as a client in YARN, spark.executor.cores set to two as the number of CPUs of the virtual machine, and spark.app.name, the name of the application. The name of the app is particularly useful when the (YARN) cluster is shared; going to ht.0.0.1:8088, it is possible to check the state of the application:
The data model used by Spark is named Resilient Distributed Dataset (RDD), which is a distributed collection of elements that can be processed in parallel. An RDD can be created from an existing collection (a Python list, for example) or from an external dataset, stored as a file on the local machine, HDFS, or other sources.
Let's now create an RDD containing integers from 0 to 9. To do so, we can use the parallelize method provided by the SparkContext object:
In:numbers = range(10) numbers_rdd = sc.parallelize(numbers) numbers_rdd Out:ParallelCollectionRDD[1] at parallelize at PythonRDD.scala:423
As you can see, you can't simply print the RDD content as it's split into multiple partitions (and distributed in the cluster). The default number of partitions is twice the number of CPUs (so, it's four in the provided VM), but it can be set manually using the second argument of the parallelize method.
To print out the data contained in the RDD, you should call the collect method. Note that this operation, while run on a cluster, collects all the data on the node; therefore, the node should have enough memory to contain it all:
In:numbers_rdd.collect() Out:[0, 1, 2, 3, 4, 5, 6, 7, 8, 9]
To obtain just a partial peek, use the take method indicating how many elements you'd want to see. Note that as it's a distributed dataset, it's not guaranteed that elements are in the same order as when we inserted it:
In:numbers_rdd.take() Out:[0, 1, 2, 3]
To read a text file, we can use the textFile method provided by the Spark Context. It allows reading both HDFS files and local files, and it splits the text on the newline characters; therefore, the first element of the RDD is the first line of the text file (using the first method). Note that if you're using a local path, all the nodes composing the cluster should access the same file through the same path:
In:sc.textFile("hdfs:///datasets/hadoop_git_readme.txt").first()
Out:u'For the latest information about Hadoop, please visit our website at:'
In:sc.textFile("file:///home/vagrant/datasets/hadoop_git_readme.txt").first()
Out:u'For the latest information about Hadoop, please visit our website at:'To save the content of an RDD on disk, you can use the saveAsTextFile method provided by the RDD. Here, you can use multiple destinations; in this example, let's save it in HDFS and then list the content of the output:
In:numbers_rdd.saveAsTextFile("hdfs:///tmp/numbers_1_10.txt")
In:!hdfs dfs -ls /tmp/numbers_1_10.txt
Out:Found 5 items
-rw-r--r-- 1 vagrant supergroup 0 2016-02-12 14:18 /tmp/numbers_1_10.txt/_SUCCESS
-rw-r--r-- 1 vagrant supergroup 4 2016-02-12 14:18 /tmp/numbers_1_10.txt/part-00000
-rw-r--r-- 1 vagrant supergroup 4 2016-02-12 14:18 /tmp/numbers_1_10.txt/part-00001
-rw-r--r-- 1 vagrant supergroup 4 2016-02-12 14:18 /tmp/numbers_1_10.txt/part-00002
-rw-r--r-- 1 vagrant supergroup 8 2016-02-12 14:18 /tmp/numbers_1_10.txt/part-00003Spark writes one file for each partition, exactly as MapReduce, writing one file for each reducer. This way speeds up the saving time as each partition is saved independently, but on a 1-node cluster, it makes things harder to read.
Can we take all the partitions to 1 before writing the file or, generically, can we lower the number of partitions in an RDD? The answer is yes, through the coalesce method provided by the RDD, passing the number of partitions we'd want to have as an argument. Passing 1 forces the RDD to be in a standalone partition and, when saved, produces just one output file. Note that this happens even when saving on the local filesystem: a file is created for each partition. Mind that doing so on a cluster environment composed by multiple nodes won't ensure that all the nodes see the same output files:
In:
numbers_rdd.coalesce(1)
.saveAsTextFile("hdfs:///tmp/numbers_1_10_one_file.txt")
In : !hdfs dfs -ls /tmp/numbers_1_10_one_file.txt
Out:Found 2 items
-rw-r--r-- 1 vagrant supergroup 0 2016-02-12 14:20 /tmp/numbers_1_10_one_file.txt/_SUCCESS
-rw-r--r-- 1 vagrant supergroup 20 2016-02-12 14:20 /tmp/numbers_1_10_one_file.txt/part-00000
In:!hdfs dfs -cat /tmp/numbers_1_10_one_file.txt/part-00000
Out:0
1
2
3
4
5
6
7
8
9
In:numbers_rdd.saveAsTextFile("file:///tmp/numbers_1_10.txt")
In:!ls /tmp/numbers_1_10.txt
Out:part-00000 part-00001 part-00002 part-00003 _SUCCESSAn RDD supports just two types of operations:
- Transformations transform the dataset into a different one. Inputs and outputs of transformations are both RDDs; therefore, it's possible to chain together multiple transformations, approaching a functional style programming. Moreover, transformations are lazy, that is, they don't compute their results straightaway.
- Actions return values from RDDs, such as the sum of the elements and the count, or just collect all the elements. Actions are the trigger to execute the chain of (lazy) transformations as an output is required.
Typical Spark programs are a chain of transformations with an action at the end. By default, all the transformations on the RDD are executed each time you run an action (that is, the intermediate state after each transformer is not saved). However, you can override this behavior using the persist method (on the RDD) whenever you want to cache the value of the transformed elements. The persist method allows both memory and disk persistency.
In the next example, we will square all the values contained in an RDD and then sum them up; this algorithm can be executed through a mapper (square elements) followed by a reducer (summing up the array). According to Spark, the map method is a transformer as it just transforms the data element by element; reduce is an action as it creates a value out of all the elements together.
Let's approach this problem step by step to see the multiple ways in which we can operate. First, start with the mapping: we first define a function that returns the square of the input argument, then we pass this function to the map method in the RDD, and finally we collect the elements in the RDD:
In:
def sq(x):
return x**2
numbers_rdd.map(sq).collect()
Out:[0, 1, 4, 9, 16, 25, 36, 49, 64, 81]Although the output is correct, the sq function is taking a lot of space; we can rewrite the transformation more concisely, thanks to Python's lambda expression, in this way:
In:numbers_rdd.map(lambda x: x**2).collect() Out:[0, 1, 4, 9, 16, 25, 36, 49, 64, 81]
Remember: why did we need to call collect to print the values in the transformed RDD? This is because the map method will not spring to action, but will be just lazily evaluated. The reduce method, on the other hand, is an action; therefore, adding the reduce step to the previous RDD should output a value. As for map, reduce takes as an argument a function that should have two arguments (left value and right value) and should return a value. Even in this case, it can be a verbose function defined with def or a lambda function:
In:numbers_rdd.map(lambda x: x**2).reduce(lambda a,b: a+b) Out:285
To make it even simpler, we can use the sum action instead of the reducer:
In:numbers_rdd.map(lambda x: x**2).sum() Out:285
So far, we've shown a very simple example of pySpark. Think about what's going on under the hood: the dataset is first loaded and partitioned across the cluster, then the mapping operation is run on the distributed environment, and then all the partitions are collapsed together to generate the result (sum or reduce), which is finally printed on the IPython Notebook. A huge task, yet made super simple by pySpark.
Let's now advance one step and introduce the key-value pairs; although RDDs can contain any kind of object (we've seen integers and lines of text so far), a few operations can be made when the elements are tuples composed by two elements: key and value.
To show an example, let's now first group the numbers in the RDD in odds and evens and then compute the sum of the two groups separately. As for the MapReduce model, it would be nice to map each number with a key (odd or even) and then, for each key, reduce using a sum operation.
We can start with the map operation: let's first create a function that tags the numbers, outputting even if the argument number is even, odd otherwise. Then, create a key-value mapping that creates a key-value pair for each number, where the key is the tag and the value is the number itself:
In:
def tag(x):
return "even" if x%2==0 else "odd"
numbers_rdd.map(lambda x: (tag(x), x) ).collect()
Out:[('even', 0),
('odd', 1),
('even', 2),
('odd', 3),
('even', 4),
('odd', 5),
('even', 6),
('odd', 7),
('even', 8),
('odd', 9)]To reduce each key separately, we can now use the reduceByKey method (which is not a Spark action). As an argument, we should pass the function that we should apply to all the values of each key; in this case, we will sum up all of them. Finally, we should call the collect method to print the results:
In:
numbers_rdd.map(lambda x: (tag(x), x) )
.reduceByKey(lambda a,b: a+b).collect()
Out:[('even', 20), ('odd', 25)]Now, let's list some of the most important methods available in Spark; it's not an exhaustive guide, but just includes the most used ones.
We start with transformations; they can be applied to an RDD and they produce an RDD:
map(function): This returns an RDD formed by passing each element through the function.flatMap(function): This returns an RDD formed by flattening the output of the function for each element of the input RDD. It's used when each value at the input can be mapped to 0 or more output elements.For example, to count the number of times that each word appears in a text, we should map each word to a key-value pair (the word would be the key,
1the value), producing more than one key-value element for each input line of text in this way:filter(function): This returns a dataset composed by all the values where the function returns true.sample(withReplacement, fraction, seed): This bootstraps the RDD, allowing you to create a sampled RDD (with or without replacement) whose length is a fraction of the input one.distinct(): This returns an RDD containing distinct elements of the input RDD.coalesce(numPartitions): This decreases the number of partitions in the RDD.repartition(numPartitions): This changes the number of partitions in the RDD. This methods always shuffles all the data over the network.groupByKey(): This creates an RDD where, for each key, the value is a sequence of values that have that key in the input dataset.reduceByKey(function): This aggregates the input RDD by key and then applies the reduce function to the values of each group.sortByKey(ascending): This sorts the elements in the RDD by key in ascending or descending order.union(otherRDD): This merges two RDDs together.intersection(otherRDD): This returns an RDD composed by just the values appearing both in the input and argument RDD.join(otherRDD): This returns a dataset where the key-value inputs are joined (on the key) to the argument RDD.Similar to the join function in SQL, there are available these methods as well:
cartesian,leftOuterJoin,rightOuterJoin, andfullOuterJoin.
Now, let's overview what are the most popular actions available in pySpark. Note that actions trigger the processing of the RDD through all the transformers in the chain:
reduce(function): This aggregates the elements of the RDD producing an output valuecount(): This returns the count of the elements in the RDDcountByKey(): This returns a Python dictionary, where each key is associated with the number of elements in the RDD with that keycollect(): This returns all the elements in the transformed RDD locallyfirst(): This returns the first value of the RDDtake(N): This returns the first N values in the RDDtakeSample(withReplacement, N, seed): This returns a bootstrap of N elements in the RDD with or without replacement, eventually using the random seed provided as argumenttakeOrdered(N, ordering): This returns the top N element in the RDD after having sorted it by value (ascending or descending)saveAsTextFile(path): This saves the RDD as a set of text files in the specified directory
There are also a few methods that are neither transformers nor actions:
cache(): This caches the elements of the RDD; therefore, future computations based on the same RDD can reuse this as a starting pointpersist(storage): This is the same as cache, but you can specify where to store the elements of RDD (memory, disk, or both)unpersist(): This undoes the persist or cache operation
Let's now try to replicate the examples that we've seen in the section about MapReduce with Hadoop. With Spark, the algorithm should be as follows:
- The input file is read and parallelized on an RDD. This operation can be done with the
textFilemethod provided by the Spark Context. - For each line of the input file, three key-value pairs are returned: one containing the number of chars, one the number of words, and the last the number of lines. In Spark, this is a flatMap operation as three outputs are generated for each input line.
- For each key, we sum up all the values. This can be done with the
reduceByKeymethod. - Finally, results are collected. In this case, we can use the
collectAsMapmethod that collects the key-value pairs in the RDD and returns a Python dictionary. Note that this is an action; therefore, the RDD chain is executed and a result is returned.
In:
def emit_feats(line):
return [("chars", len(line)),
("words", len(line.split())),
("lines", 1)]
print (sc.textFile("/datasets/hadoop_git_readme.txt")
.flatMap(emit_feats)
.reduceByKey(lambda a,b: a+b)
.collectAsMap())
Out:{'chars': 1335, 'lines': 31, 'words': 179}We can immediately note the enormous speed of this method compared to the MapReduce implementation. This is because all of the dataset is stored in-memory and not in HDFS. Secondly, this is a pure Python implementation and we don't need to call external command lines or libraries—pySpark is self-contained.
Let's now work on the example on the larger file, containing the Shakespeare texts, to extract the most popular word. In the Hadoop MapReduce implementation, it takes two map-reduce steps and therefore four write/read on HDFS. In pySpark, we can do all this in an RDD:
- The input file is read and parallelized on an RDD with the
textFilemethod. - For each line, all the words are extracted. For this operation, we can use the flatMap method and a regular expression.
- Each word in the text (that is, each element of the RDD) is now mapped to a key-value pair: the key is the lowercased word and the value is always
1. This is a map operation. - With a
reduceByKeycall, we count how many times each word (key) appears in the text (RDD). The output is key-value pairs, where the key is a word and value is the number of times the word appears in the text. - We flip keys and values, creating a new RDD. This is a map operation.
- We sort the RDD in descending order and extract (take) the first element. This is an action and can be done in one operation with the
takeOrderedmethod.
In:import re
WORD_RE = re.compile(r"[w']+")
print (sc.textFile("/datasets/shakespeare_all.txt")
.flatMap(lambda line: WORD_RE.findall(line))
.map(lambda word: (word.lower(), 1))
.reduceByKey(lambda a,b: a+b)
.map(lambda (k,v): (v,k))
.takeOrdered(1, key = lambda x: -x[0]))
Out:[(27801, u'the')]The results are the same that we had using Hadoop and MapReduce, but in this case, the computation takes far less time. We can actually further improve the solution, collapsing the second and third steps together (flatMap-ing a key-value pair for each word, where the key is the lowercased word and value is the number of occurrences) and the fifth and sixth steps together (taking the first element and ordering the elements in the RDD by their value, that is, the second element of the pair):
In:
print (sc.textFile("/datasets/shakespeare_all.txt")
.flatMap(lambda line: [(word.lower(), 1) for word in WORD_RE.findall(line)])
.reduceByKey(lambda a,b: a+b)
.takeOrdered(1, key = lambda x: -x[1]))
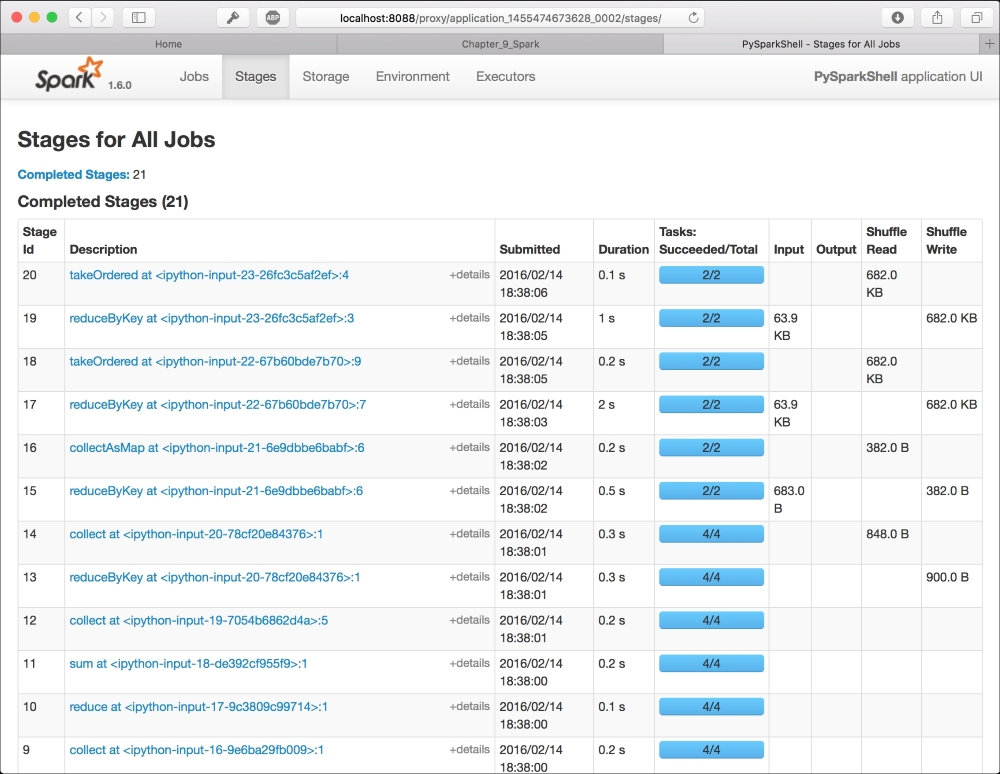
Out:[(u'the', 27801)]To check the state of the processing, you can use the Spark UI: it's a graphical interface that shows the jobs run by Spark step-by-step. To access the UI, you should first figure out what's the name of the pySpark IPython application, searching in the bash shell where you've launched the notebook by its name (typically, it is in the form application_<number>_<number>), and then point your browser to the page: http://localhost:8088/proxy/application_<number>_<number>
The result is similar to the one in the following image. It contains all the jobs run in spark (as IPython Notebook cells), and you can also visualize the execution plan as a directed acyclic graph (DAG):