Principal Components Analysis (PCA) is a dimensionality reduction technique that's used very frequently in computer vision and machine learning. When we deal with features with large dimensionalities, training a machine learning system becomes prohibitively expensive. Therefore, we need to reduce the dimensionality of the data before we can train a system. However, when we reduce the dimensionality, we don't want to lose the information present in the data. This is where PCA comes into the picture! PCA identifies the important components of the data and arranges them in the order of importance. You can learn more about it at http://dai.fmph.uniba.sk/courses/ml/sl/PCA.pdf. It is used a lot in face recognition systems. Let's see how to perform PCA on input data.
- Create a new Python file, and import the following packages:
import numpy as np from sklearn import decomposition
- Let's define five dimensions for our input data. The first two dimensions will be independent, but the next three dimensions will be dependent on the first two dimensions. This basically means that we can live without the last three dimensions because they do not give us any new information:
# Define individual features x1 = np.random.normal(size=250) x2 = np.random.normal(size=250) x3 = 2*x1 + 3*x2 x4 = 4*x1 - x2 x5 = x3 + 2*x4
- Let's create a dataset with these features.
# Create dataset with the above features X = np.c_[x1, x3, x2, x5, x4]
- Create a PCA object:
# Perform Principal Components Analysis pca = decomposition.PCA()
- Fit a PCA model on the input data:
pca.fit(X)
- Print the variances of the dimensions:
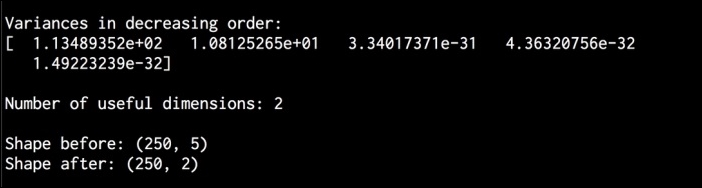
# Print variances variances = pca.explained_variance_ print ' Variances in decreasing order: ', variances
- If a particular dimension is useful, then it will have a meaningful value for the variance. Let's set a threshold and identify the important dimensions:
# Find the number of useful dimensions thresh_variance = 0.8 num_useful_dims = len(np.where(variances > thresh_variance)[0]) print ' Number of useful dimensions:', num_useful_dims
- Just like we discussed earlier, PCA identified that only two dimensions are important in this dataset:
# As we can see, only the 2 first components are useful pca.n_components = num_useful_dims
- Let's convert the dataset from a five-dimensional set to a two-dimensional set:
X_new = pca.fit_transform(X) print ' Shape before:', X.shape print 'Shape after:', X_new.shape
- The full code is given in the
pca.pyfile that's already provided to you for reference. If you run this code, you will see the following on your Terminal: